Chaperone-mediated autophagy
Chaperone-mediated autophagy (CMA) refers to the chaperone-dependent selection of soluble cytosolic proteins that are then targeted to lysosomes and directly translocated across the lysosome membrane for degradation.[1] The unique features of this type of autophagy are the selectivity on the proteins that are degraded by this pathway and the direct shuttling of these proteins across the lysosomal membrane without the requirement for the formation of additional vesicles (Figure 1).

Molecular components and steps of CMA
The proteins that are degraded through CMA are cytosolic proteins or proteins from other compartments once they reach the cytosol. Therefore, some of the components that participate in CMA are present in the cytosol while others are located at the lysosomal membrane (Table I).
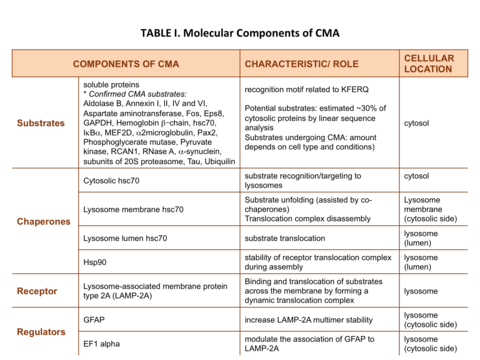
For a protein to be a CMA substrate, it must have in its amino acid sequence a pentapeptide motif biochemically related to KFERQ.[2] This CMA-targeting motif is recognized by a cytosolic chaperone, heat shock cognate protein of 70 kDa (hsc70) which targets the substrate to the lysosome surface.[3] This substrate protein-chaperone complex binds to lysosome-associated membrane protein type 2A (LAMP-2A), which acts as the receptor for this pathway.[4] LAMP-2A a single span membrane protein, is one of the three spliced variants of a single gene lamp2.[5] The other two isoforms LAMP-2B and LAMP-2C are involved in macroautophagy and vesicular trafficking, respectively. Substrate proteins undergo unfolding after binding to LAMP-2A in a process likely mediated by the membrane associated hsc70 and its co-chaperones Bag1, hip, hop and hsp40, also detected at the lysosomal membrane.[6] This binding of substrates to monomers of LAMP-2A triggers the assembly of LAMP-2A multimers that act as the active translocation complex through which the substrates can pass through after unfolding.[7] Substrate translocation requires the presence of hsc70 inside the lysosomal lumen, which may act by either pulling substrates into the lysosomes or preventing their return to the cytosol.[8] After translocation the substrate proteins are rapidly degraded by the lysosomal proteases. Figure 1 depicts the different steps of CMA.
The limiting step for CMA is the binding of the substrate proteins to LAMP-2A and, consequently, levels of LAMP-2A at the lysosomal membrane correlate directly with CMA activity. Therefore, to modulate the activity of this autophagic pathway, the cell stringently regulates the levels of the CMA receptor at the lysosomal membrane by controlling the degradation rates of LAMP-2A monomers in lysosomes and by de novo synthesis of LAMP-2A molecules. In addition, transport of substrates also depends on the efficiency of the assembly of LAMP-2A into the translocation complex.[7]
Assembly and disassembly of CMA translocation complex is mediated by hsp90 and hsc70 chaperones, respectively.[7] Degradation of LAMP-2A monomers at the lysosomal membrane occurs in discrete cholesterol-rich lipid microdomains of the lysosomal membrane and it is mediated by Cathepsin A and an unidentified lysosomal metalloprotease.[9] Therefore, assembly, disassembly of LAMP-2A into active translocation complex, and its degradation in microdomain regions, highlights the dynamic nature of this process and the importance of lateral mobility of the CMA receptor at the lysosomal membrane.
Physiological functions of CMA
CMA contributes to the maintenance of cellular homeostasis by facilitating recycling of amino acids of the degraded proteins (contribution to energetic cellular balance) and by eliminating abnormal or damaged proteins (contribution to cellular quality control).[1]
CMA is active at all times in different tissues (liver, kidney, brain), and almost all cell types in culture studied. However, it is maximally activated in response to stressors and changes in the cellular nutritional status. When nutrient supply is limited, the cells respond by activating autophagy, in order to degrade intracellular components to provide energy and building blocks, which the cell can utilize in this dire state.[10] Macroautophagy is activated as early as 30 minutes into starvation and remains at high activity for at least 4–8 hours into starvation. If the starvation state persists for more than 10 hours, the cells switch to the selective form of autophagy, namely CMA, which is known to reach a plateau of maximal activation ~36 hours into fasting and remains at these levels until ~3 days. The selectivity of CMA for individual cytosolic proteins permits cells to degrade only those proteins that might not be required in these starvation conditions in order to generate amino acids for the synthesis of essential proteins. For example, some of the best-characterized CMA substrates are enzymes involved in glycolysis, a pathway known to be less active in fasting conditions.[11][12]
CMA is important in regulating cellular metabolism. Specific depletion of CMA in liver results in robust hepatic glycogen use accompanied with accumulation of fat in the liver, along with altered glucose homeostasis, increased energy expenditure and reduced peripheral adiposity.[12] Proteomics analyses identified several enzymes of the carbohydrate and the lipid metabolism pathways to be CMA substrates, and their altered degradation in the knockout mice explaining the abnormal metabolic phenotype of the CMA-deficient mice.[12]
CMA activity has been shown to be modulated through retinoic acid receptor alpha signaling and is specifically activated by designed all-trans retinoic acid derivatives in cultured cells.[13]
CMA is also responsible for the selective removal of damaged and no-longer-functional proteins. This function is critical when cells are exposed to agents that cause protein damage as the selectivity of CMA ensures that only the damaged proteins get targeted to lysosomes for degradation. For instance, oxidative stress and exposure to toxic compounds are stimuli that upregulate CMA.[14] Consequently, cells that are defective for CMA are more susceptible to these insults than control cells.[15]
CMA performs various specialized functions as well, depending on the specific protein undergoing degradation through this pathway and the cell type involved. For example, known CMA substrates include, MEF2D, a neuronal factor important for survival; Pax2, a transcription factor, important for the regulation growth of renal tubular cells; IκBα, known inhibitor of NFκB. CMA has also been suggested to contribute to antigen presentation in dendritic cells.[16][17][18]
CMA is activated during T cell activation due to increased expression of the CMA receptor LAMP-2A.[19] CMA is essential for T cell activation through the degradation of negative regulators of T cell activation (Itch, RCAN1). Consequently, specific depletion of CMA in T cells results in immune response deficiency following immunization or infection.[19]
CMA is increased upon genotoxic stress.[20] Conversely, decreased CMA activity associates with increased genome instability and decreased cell survival. CMA is involved in the removal of Chk1, a key protein for cell cycle progression and cells with impaired CMA have defective DNA repair.[20]
CMA degrades lipid droplet proteins (perilipin 2 and perilipin 3).[21] Removal of these lipid droplet coat proteins by CMA precedes lipolysis and lipophagy.[21] Consequently, defective CMA activity leads to massive accumulation of lipid droplets and steatosis.[12][21]
Pathology of CMA
CMA activity declines with age in many cell types of old rodents and in cells of older humans.[22][23][24] This impairment of CMA in aging is mainly due to a decrease in the levels of LAMP-2A at the lysosomal membrane, because of reduced stability of the CMA receptor and not due to decreased de novo synthesis. Studies in a transgenic mouse model in which normal levels of LAMP-2A are maintained throughout life, showed that these animals had ‘cleaner’ cells, better response to stress – and overall, a better health-span.[24] These studies support the possible contribution of declined CMA activity to poor cellular homeostasis and inefficient response to stress characteristic of old organisms. High-fat diet inhibits CMA.[25] This is because of a decrease in the stability of the CMA receptor at the lysosomal surface.
A primary defect in CMA activity has also been described in neurodegenerative diseases, such as Parkinson’s disease[26][27][28] and certain tauopathies.[29] In these cases, the defect lies in the ‘tight’ binding to the lysosomal membrane of pathogenic proteins known to accumulate in these disorders (α-synuclein, UCHL1 in Parkinson’s disease and mutant Tau in tauopathies). These toxic proteins often bind to LAMP-2A with abnormal affinity exerting a ‘clogging effect’ at the lysosomal membrane and thus, inhibit the CMA-mediated degradation of other cytosolic substrate proteins.[26][27]
Links between CMA and cancer have also been established.[30][31][32] CMA is upregulated in many different types of human cancer cells and blockage of CMA in these cells reduces their proliferative, tumorigenic and metastatic capabilities. In fact, interference of the expression of LAMP-2A in already-formed experimental tumors in mice resulted in their regression.[30]
References
- 1 2 Kaushik, Susmita; Cuervo, Ana Maria (2012). "Chaperone-mediated autophagy: A unique way to enter the lysosome world". Trends in Cell Biology. 22 (8): 407–17. doi:10.1016/j.tcb.2012.05.006. PMC 3408550
 . PMID 22748206.
. PMID 22748206. - ↑ Fred Dice, J. (1990). "Peptide sequences that target cytosolic proteins for lysosomal proteolysis". Trends in Biochemical Sciences. 15 (8): 305–9. doi:10.1016/0968-0004(90)90019-8. PMID 2204156.
- ↑ Chiang, H.; Terlecky, SR; Plant, C.; Dice, J. (1989). "A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins". Science. 246 (4928): 382–5. Bibcode:1989Sci...246..382C. doi:10.1126/science.2799391. PMID 2799391.
- ↑ Cuervo, A. M.; Dice, J. F. (1996). "A Receptor for the Selective Uptake and Degradation of Proteins by Lysosomes". Science. 273 (5274): 501–3. Bibcode:1996Sci...273..501C. doi:10.1126/science.273.5274.501. PMID 8662539.
- ↑ Eskelinen, Eeva-Liisa; Cuervo, Ana Maria; Taylor, Matthew R.G.; Nishino, Ichizo; Blum, Janice S.; Dice, J. Fred; Sandoval, Ignacio V.; Lippincott-Schwartz, Jennifer; et al. (2005). "Unifying Nomenclature for the Isoforms of the Lysosomal Membrane Protein LAMP-2". Traffic. 6 (11): 1058–61. doi:10.1111/j.1600-0854.2005.00337.x. PMID 16190986.
- ↑ Salvador, N. (2000). "Import of a Cytosolic Protein into Lysosomes by Chaperone-Mediated Autophagy depends on its Folding State". Journal of Biological Chemistry. doi:10.1074/jbc.M001394200.
- 1 2 3 Bandyopadhyay, U.; Kaushik, S.; Varticovski, L.; Cuervo, A. M. (2008). "The Chaperone-Mediated Autophagy Receptor Organizes in Dynamic Protein Complexes at the Lysosomal Membrane". Molecular and Cellular Biology. 28 (18): 5747–63. doi:10.1128/MCB.02070-07. PMC 2546938
 . PMID 18644871.
. PMID 18644871. - ↑ Agarraberes, F. A.; Terlecky, SR; Dice, JF (1997). "An Intralysosomal hsp70 is Required for a Selective Pathway of Lysosomal Protein Degradation". The Journal of Cell Biology. 137 (4): 825–34. doi:10.1083/jcb.137.4.825. PMC 2139836
 . PMID 9151685.
. PMID 9151685. - ↑ Kaushik, Susmita; Massey, Ashish C; Cuervo, Ana Maria (2006). "Lysosome membrane lipid microdomains: Novel regulators of chaperone-mediated autophagy". The EMBO Journal. 25 (17): 3921–33. doi:10.1038/sj.emboj.7601283. PMC 1560360
 . PMID 16917501.
. PMID 16917501. - ↑ Cuervo, AM; Knecht, E; Terlecky, SR; Dice, JF (1995). "Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation". The American journal of physiology. 269 (5 Pt 1): C1200–8. PMID 7491910.
- ↑ Aniento, F; Roche, E; Cuervo, AM; Knecht, E (1993). "Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes". The Journal of Biological Chemistry. 268 (14): 10463–70. PMID 8486700.
- 1 2 3 4 Schneider, JL; Suh, Y; Cuervo, AM (2 September 2014). "Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation.". Cell Metabolism. 20 (3): 417–32. doi:10.1016/j.cmet.2014.06.009. PMID 25043815.
- ↑ Anguiano, J; Garner, TP; Mahalingam, M; Das, BC; Gavathiotis, E; Cuervo, AM (June 2013). "Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives.". Nature Chemical Biology. 9 (6): 374–82. doi:10.1038/nchembio.1230. PMID 23584676.
- ↑ Kiffin, R.; Christian, C; Knecht, E; Cuervo, AM (2004). "Activation of Chaperone-mediated Autophagy during Oxidative Stress". Molecular Biology of the Cell. 15 (11): 4829–40. doi:10.1091/mbc.E04-06-0477. PMC 524731
 . PMID 15331765.
. PMID 15331765. - ↑ Massey, A. C.; Kaushik, S.; Sovak, G.; Kiffin, R.; Cuervo, A. M. (2006). "Consequences of the selective blockage of chaperone-mediated autophagy". Proceedings of the National Academy of Sciences. 103 (15): 5805–5810. Bibcode:2006PNAS..103.5805M. doi:10.1073/pnas.0507436103.
- ↑ Yang, Q.; She, H.; Gearing, M.; Colla, E.; Lee, M.; Shacka, J. J.; Mao, Z. (2009). "Regulation of Neuronal Survival Factor MEF2D by Chaperone-Mediated Autophagy". Science. 323 (5910): 124–7. Bibcode:2009Sci...323..124Y. doi:10.1126/science.1166088. PMC 2666000
 . PMID 19119233.
. PMID 19119233. - ↑ Zhou, Delu; Li, Ping; Lin, Yinling; Lott, Jeremy M.; Hislop, Andrew D.; Canaday, David H.; Brutkiewicz, Randy R.; Blum, Janice S. (2005). "Lamp-2a Facilitates MHC Class II Presentation of Cytoplasmic Antigens". Immunity. 22 (5): 571–81. doi:10.1016/j.immuni.2005.03.009. PMID 15894275.
- ↑ Sooparb, Sira; Price, S. Russ; Shaoguang, Jin; Franch, Harold A. (2004). "Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus". Kidney International. 65 (6): 2135–44. doi:10.1111/j.1523-1755.2004.00639.x. PMID 15149326.
- 1 2 Valdor, R; Mocholi, E; Botbol, Y; Guerrero-Ros, I; Chandra, D; Koga, H; Gravekamp, C; Cuervo, AM; Macian, F (November 2014). "Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation.". Nature Immunology. 15 (11): 1046–54. doi:10.1038/ni.3003. PMID 25263126.
- 1 2 Park, Caroline (2015). "Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage.". Nature Communications. 6: 6823. doi:10.1038/ncomms7823. PMID 25880015.
- 1 2 3 Kaushik, Susmita (2015). "Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis". Nature Cell Biology. 17 (6): 759–70. doi:10.1038/ncb3166. PMID 25961502.
- ↑ Cuervo, A. M.; Dice, JF (2000). "Age-related Decline in Chaperone-mediated Autophagy". Journal of Biological Chemistry. 275 (40): 31505–13. doi:10.1074/jbc.M002102200. PMID 10806201.
- ↑ Kiffin, R.; Kaushik, S.; Zeng, M.; Bandyopadhyay, U.; Zhang, C.; Massey, A. C.; Martinez-Vicente, M.; Cuervo, A. M. (2007). "Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age". Journal of Cell Science. 120 (5): 782–91. doi:10.1242/jcs.001073. PMID 17284523.
- 1 2 Zhang, Cong; Cuervo, Ana Maria (2008). "Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function". Nature Medicine. 14 (9): 959–65. doi:10.1038/nm.1851. PMC 2722716
 . PMID 18690243.
. PMID 18690243. - ↑ Rodriguez-Navarro, JA; Kaushik, S; Koga, H; Dall'Armi, C; Shui, G; Wenk, MR; Di Paolo, G; Cuervo, AM (20 March 2012). "Inhibitory effect of dietary lipids on chaperone-mediated autophagy.". Proceedings of the National Academy of Sciences of the United States of America. 109 (12): E705–14. Bibcode:2012PNAS..109E.705R. doi:10.1073/pnas.1113036109. PMID 22331875.
- 1 2 Cuervo, A. M.; Stefanis, L; Fredenburg, R; Lansbury, PT; Sulzer, D (2004). "Impaired Degradation of Mutant -Synuclein by Chaperone-Mediated Autophagy". Science. 305 (5688): 1292–5. Bibcode:2004Sci...305.1292C. doi:10.1126/science.1101738. PMID 15333840.
- 1 2 Martinez-Vicente, Marta; Talloczy, Zsolt; Kaushik, Susmita; Massey, Ashish C.; Mazzulli, Joseph; Mosharov, Eugene V.; Hodara, Roberto; Fredenburg, Ross; et al. (2008). "Dopamine-modified α-synuclein blocks chaperone-mediated autophagy". Journal of Clinical Investigation. 118 (2): 777–88. doi:10.1172/JCI32806. PMC 2157565
 . PMID 18172548.
. PMID 18172548. - ↑ Orenstein, SJ; Kuo, SH; Tasset, I; Arias, E; Koga, H; Fernandez-Carasa, I; Cortes, E; Honig, LS; Dauer, W; Consiglio, A; Raya, A; Sulzer, D; Cuervo, AM (April 2013). "Interplay of LRRK2 with chaperone-mediated autophagy.". Nature Neuroscience. 16 (4): 394–406. doi:10.1038/nn.3350. PMID 23455607.
- ↑ Wang, Y.; Martinez-Vicente, M.; Kruger, U.; Kaushik, S.; Wong, E.; Mandelkow, E.-M.; Cuervo, A. M.; Mandelkow, E. (2009). "Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing". Human Molecular Genetics. 18 (21): 4153–70. doi:10.1093/hmg/ddp367. PMC 2758146
 . PMID 19654187.
. PMID 19654187. - 1 2 Kon, M.; Kiffin, R.; Koga, H.; Chapochnick, J.; MacIan, F.; Varticovski, L.; Cuervo, A. M. (2011). "Chaperone-Mediated Autophagy is Required for Tumor Growth". Science Translational Medicine. 3 (109): 109ra117. doi:10.1126/scitranslmed.3003182. PMID 22089453.
- ↑ Lv, Lei; Li, Dong; Zhao, Di; Lin, Ruiting; Chu, Yajing; Zhang, Heng; Zha, Zhengyu; Liu, Ying; et al. (2011). "Acetylation Targets the M2 Isoform of Pyruvate Kinase for Degradation through Chaperone-Mediated Autophagy and Promotes Tumor Growth". Molecular Cell. 42 (6): 719–30. doi:10.1016/j.molcel.2011.04.025. PMID 21700219.
- ↑ Quintavalle, C; Di Costanzo, S; Zanca, C; Tasset, I; Fraldi, A; Incoronato, M; Mirabelli, P; Monti, M; Ballabio, A; Pucci, P; Cuervo, AM; Condorelli, G (October 2014). "Phosphorylation-regulated degradation of the tumor-suppressor form of PED by chaperone-mediated autophagy in lung cancer cells.". Journal of cellular physiology. 229 (10): 1359–68. doi:10.1002/jcp.24569. PMID 24477641.
Further reading
- Mizushima, N; Levine, B; Cuervo, AM; Klionsky, DJ (28 February 2008). "Autophagy fights disease through cellular self-digestion.". Nature. 451 (7182): 1069–75. Bibcode:2008Natur.451.1069M. doi:10.1038/nature06639. PMC 2670399
 . PMID 18305538.
. PMID 18305538. - Kaushik, S; Cuervo, AM (August 2012). "Chaperone-mediated autophagy: a unique way to enter the lysosome world.". Trends in Cell Biology. 22 (8): 407–17. doi:10.1016/j.tcb.2012.05.006. PMC 3408550
 . PMID 22748206.
. PMID 22748206. - Arias, E; Cuervo, AM (April 2011). "Chaperone-mediated autophagy in protein quality control.". Current opinion in cell biology. 23 (2): 184–9. doi:10.1016/j.ceb.2010.10.009. PMC 3078170
 . PMID 21094035.
. PMID 21094035. - Cuervo, AM; Wong, E (January 2014). "Chaperone-mediated autophagy: roles in disease and aging.". Cell research. 24 (1): 92–104. doi:10.1038/cr.2013.153. PMID 24281265.
- Kaushik, S; Bandyopadhyay, U; Sridhar, S; Kiffin, R; Martinez-Vicente, M; Kon, M; Orenstein, SJ; Wong, E; Cuervo, AM (15 February 2011). "Chaperone-mediated autophagy at a glance.". Journal of Cell Science. 124 (Pt 4): 495–9. doi:10.1242/jcs.073874. PMID 21282471.
- Cuervo, AM (13 July 2011). "Chaperone-mediated autophagy: Dice's 'wild' idea about lysosomal selectivity.". Nature reviews. Molecular cell biology. 12 (8): 535–41. doi:10.1038/nrm3150. PMID 21750569.
- Kaushik, S; Cuervo, AM (2009). "Methods to monitor chaperone-mediated autophagy.". Methods in enzymology. 452: 297–324. doi:10.1016/s0076-6879(08)03619-7. PMID 19200890.