Clique problem
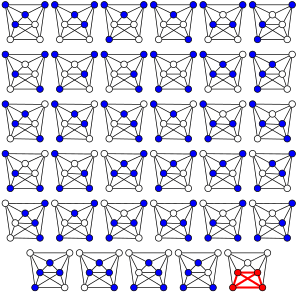
In computer science, the clique problem is the computational problem of finding cliques (subsets of vertices, all adjacent to each other, also called complete subgraphs) in a graph. It has several different formulations depending on which cliques, and what information about the cliques, should be found. Common formulations of the clique problem include finding a maximum clique (a clique with the largest possible number of vertices), finding a maximum weight clique in a weighted graph, listing all maximal cliques (cliques that cannot be enlarged), and solving the decision problem of testing whether a graph contains a clique larger than a given size.
The clique problem arises in the following real-world setting. Consider a social network, where the graph's vertices represent people, and the graph's edges represent mutual acquaintance. Then a clique represents a subset of people who all know each other, and algorithms for finding cliques can be used to discover these groups of mutual friends. Along with its applications in social networks, the clique problem also has many applications in bioinformatics and computational chemistry.
Most versions of the clique problem are hard. The clique decision problem is NP-complete (one of Karp's 21 NP-complete problems). The problem of finding the maximum clique is both fixed-parameter intractable and hard to approximate. And, listing all maximal cliques may require exponential time as there exist graphs with exponentially many maximal cliques. Therefore, much of the theory about the clique problem is devoted to identifying special types of graph that admit more efficient algorithms, or to establishing the computational difficulty of the general problem in various models of computation.
To find a maximum clique, one can systematically inspect all subsets, but this sort of brute-force search is too time-consuming to be practical for networks comprising more than a few dozen vertices. Although no polynomial time algorithm is known for this problem, more efficient algorithms than the brute-force search are known. For instance, the Bron–Kerbosch algorithm can be used to list all maximal cliques in worst-case optimal time, and it is also possible to list them in polynomial time per clique.
History and applications
The study of complete subgraphs in mathematics predates the "clique" terminology. For instance, complete subgraphs make an early appearance in the mathematical literature in the graph-theoretic reformulation of Ramsey theory by Erdős & Szekeres (1935). But the term "clique" and the problem of algorithmically listing cliques both come from the social sciences, where complete subgraphs are used to model social cliques, groups of people who all know each other. Luce & Perry (1949) used graphs to model social networks, and adapted the social science terminology to graph theory. They were the first to call complete subgraphs "cliques". The first algorithm for solving the clique problem is that of Harary & Ross (1957),[1] who were motivated by the sociological application. Social science researchers have also defined various other types of cliques and maximal cliques in social network, "cohesive subgroups" of people or actors in the network all of whom share one of several different kinds of connectivity relation. Many of these generalized notions of cliques can also be found by constructing an undirected graph whose edges represent related pairs of actors from the social network, and then applying an algorithm for the clique problem to this graph.[2]
Since the work of Harary and Ross, many others have devised algorithms for various versions of the clique problem.[1] In the 1970s, researchers began studying these algorithms from the point of view of worst-case analysis. See, for instance, Tarjan & Trojanowski (1977), an early work on the worst-case complexity of the maximum clique problem. Also in the 1970s, beginning with the work of Cook (1971) and Karp (1972), researchers began using the theory of NP-completeness and related intractability results to provide a mathematical explanation for the perceived difficulty of the clique problem. In the 1990s, a breakthrough series of papers beginning with Feige et al. (1991) and reported at the time in major newspapers,[3] showed that (assuming P ≠ NP) it is not even possible to approximate the problem accurately and efficiently.
Clique-finding algorithms have been used in chemistry, to find chemicals that match a target structure[4] and to model molecular docking and the binding sites of chemical reactions.[5] They can also be used to find similar structures within different molecules.[6] In these applications, one forms a graph in which each vertex represents a matched pair of atoms, one from each of two molecules. Two vertices are connected by an edge if the matches that they represent are compatible with each other. Being compatible may mean, for instance, that the distances between the atoms within the two molecules are approximately equal, to within some given tolerance. A clique in this graph represents a set of matched pairs of atoms in which all the matches are compatible with each other.[7] A special case of this method is the use of the modular product of graphs to reduce the problem of finding the maximum common induced subgraph of two graphs to the problem of finding a maximum clique in their product.[8]
In automatic test pattern generation, finding cliques can help to bound the size of a test set.[9] In bioinformatics, clique-finding algorithms have been used to infer evolutionary trees,[10] predict protein structures,[11] and find closely interacting clusters of proteins.[12] Listing the cliques in a dependency graph is an important step in the analysis of certain random processes.[13] In mathematics, Keller's conjecture on face-to-face tiling of hypercubes was disproved by Lagarias & Shor (1992), who used a clique-finding algorithm on an associated graph to find a counterexample.[14]
Definitions
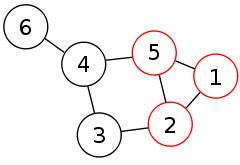
An undirected graph is formed by a finite set of vertices and a set of unordered pairs of vertices, which are called edges. By convention, in algorithm analysis, the number of vertices in the graph is denoted by n and the number of edges is denoted by m. A clique in a graph G is a complete subgraph of G. That is, it is a subset K of the vertices such that every two vertices in K are the two endpoints of an edge in G. A maximal clique is a clique to which no more vertices can be added. For each vertex v that is not part of a maximal clique, there must be another vertex w that is in the clique and non-adjacent to v, preventing v from being added to the clique. A maximum clique is a clique that includes the largest possible number of vertices. The clique number ω(G) is the number of vertices in a maximum clique of G.[1]
Several closely related clique-finding problems have been studied.[15]
- In the maximum clique problem, the input is an undirected graph, and the output is a maximum clique in the graph. If there are multiple maximum cliques, one of them may be chosen arbitrarily.[15]
- In the weighted maximum clique problem, the input is an undirected graph with weights on its vertices (or, less frequently, edges) and the output is a clique with maximum total weight. The maximum clique problem is the special case in which all weights are equal.[16] As well as the problem of optimizing the sum of weights, other more complicated bicriterion optimization problems have also been studied.[17]
- In the maximal clique listing problem, the input is an undirected graph, and the output is a list of all its maximal cliques. The maximum clique problem may be solved using as a subroutine an algorithm for the maximal clique listing problem, because the maximum clique must be included among all the maximal cliques.[18]
- In the k-clique problem, the input is an undirected graph and a number k. The output is a clique withk vertices, if one exists, or a special value indicating that there is no k-clique otherwise.
In some variations of this problem, the output should list all cliques of size k.[19]
- In the clique decision problem, the input is an undirected graph and a number k, and the output is a Boolean value: true if the graph contains a k-clique, and false otherwise.[20]
The first four of these problems are all important in practical applications. The clique decision problem is not of practical importance; it is formulated in this way in order to apply the theory of NP-completeness to clique-finding problems.[20]
The clique problem and the independent set problem are complementary: a clique in G is an independent set in the complement graph of G and vice versa.[21] Therefore, many computational results may be applied equally well to either problem, and some research papers do not clearly distinguish between the two problems. However, the two problems have different properties when applied to restricted families of graphs. For instance, the clique problem may be solved in polynomial time for planar graphs[22] while the independent set problem remains NP-hard on planar graphs.[23]
Algorithms
Finding a single maximal clique
A maximal clique, sometimes called inclusion-maximal, is a clique that is not included in a larger clique. Therefore, every clique is contained in a maximal clique.[24] Maximal cliques can be very small. A graph may contain a non-maximal clique with many vertices and a separate clique of size 2 which is maximal. While a maximum (i.e., largest) clique is necessarily maximal, the converse does not hold. There are some types of graphs in which every maximal clique is maximum; these are the complements of the well-covered graphs, in which every maximal independent set is maximum.[25] However, other graphs have maximal cliques that are not maximum.
Finding a single maximal clique is straightforward: Starting with an arbitrary clique (for instance, a single vertex), grow the current clique one vertex at a time by iterating over the graph's remaining vertices, adding a vertex if it is connected to each vertex in the current clique, and discarding it otherwise. This algorithm runs in linear time.[26] Because of the ease of finding maximal cliques, and their potential small size, more attention has been given to the much harder algorithmic problem of finding a maximum or otherwise large clique than has been given to the problem of finding a single maximal clique. However, some research in parallel algorithms has studied the problem of finding a maximal clique. In particular, the problem of finding the lexicographically first maximal clique (the one found by the greedy algorithm described above) has been shown to be complete for the class of polynomial-time functions. This result implies that the problem is unlikely to be solvable within the parallel complexity class NC.[27]
Cliques of fixed size
One can test whether a graph G contains a k-vertex clique, and find any such clique that it contains, using a brute force algorithm. This algorithm examines each subgraph with k vertices and checks to see whether it forms a clique. It takes time O(nk k2): there are O(nk) subgraphs to check, each of which has O(k2) edges whose presence in G needs to be checked. Thus, the problem may be solved in polynomial time whenever k is a fixed constant. When k is part of the input to the problem, however, the time is exponential.[28]
The simplest nontrivial case of the clique-finding problem is finding a triangle in a graph, or equivalently determining whether the graph is triangle-free. In a graph with m edges, there may be at most Θ(m3/2) triangles. The worst case for this formula occurs when G is itself a clique. Therefore, algorithms for listing all triangles must take at least Ω(m3/2) time in the worst case, and algorithms are known that match this time bound.[29] For instance, Chiba & Nishizeki (1985) describe an algorithm that sorts the vertices in order from highest degree to lowest and then iterates through each vertex v in the sorted list, looking for triangles that include v and do not include any previous vertex in the list. To do so the algorithm marks all neighbors of v, searches through all edges incident to a neighbor of v outputting a triangle for every edge that has two marked endpoints, and then removes the marks and deletes v from the graph. As the authors show, the time for this algorithm is proportional to the arboricity of the graph (denoted a(G)) multiplied by the number of edges, which is O(m a(G)). Since the arboricity is at most O(m1/2), this algorithm runs in time O(m3/2). More generally, all k-vertex cliques can be listed by a similar algorithm that takes time proportional to the number of edges multiplied by the arboricity to the power (k − 2). For graphs of constant arboricity, such as planar graphs (or in general graphs from any non-trivial minor-closed graph family), this algorithm takes O(m) time, which is optimal since it is linear in the size of the input.[19]
If one desires only a single triangle, or an assurance that the graph is triangle-free, faster algorithms are possible. As Itai & Rodeh (1978) observe, the graph contains a triangle if and only if its adjacency matrix and the square of the adjacency matrix contain nonzero entries in the same cell. Therefore, fast matrix multiplication techniques such as the Coppersmith–Winograd algorithm can be applied to find triangles in time O(n2.376). Alon, Yuster & Zwick (1994) used fast matrix multiplication to improve the O(m3/2) algorithm for finding triangles to O(m1.41). These algorithms based on fast matrix multiplicationhave also been extended to problems of finding k-cliques for larger values of k.[30]
Listing all maximal cliques
By a result of Moon & Moser (1965), every n-vertex graph has at most 3n/3 maximal cliques. They can be listed by the Bron–Kerbosch algorithm, a recursive backtracking procedure of Bron & Kerbosch (1973). The main recursive subroutine of this procedure has three arguments: a partially constructed (non-maximal) clique, a set of candidate vertices that could be added to the clique, and another set of vertices that should not be added (because doing so would lead to a clique that has already been found). The algorithm tries adding the candidate vertices one by one to the partial clique, making a recursive call for each one. After trying each of these vertices, it moves it to the set of vertices that should not be added again. Variants of this algorithm can be shown to have worst-case running time O(3n/3).[31] Therefore, this provides a worst-case-optimal solution to the problem of listing all maximal cliques. Further, the Bron–Kerbosch algorithm has been widely reported as being faster in practice than its alternatives.[32]
However, when the number of cliques is significantly smaller than its worst case, other algorithms might be preferable. As Tsukiyama et al. (1977) showed, it is also possible to list all maximal cliques in a graph in an amount of time that is polynomial per generated clique. An algorithm such as theirs in which the running time depends on the output size is known as an output-sensitive algorithm. Their algorithm is based on the following two observations, relating the maximal cliques of the given graph G to the maximal cliques of a graph G \ v formed by removing an arbitrary vertex v fromG:
- For every maximal clique K of G \ v, either K continues to form a maximal clique in G, or K ⋃ {v} forms a maximal clique in G. Therefore, G has at least as many maximal cliques as G \ v does.
- Each maximal clique in G that does not contain v is a maximal clique in G \ v, and each maximal clique in G that does contain v can be formed from a maximal clique K in G \ v by adding v and removing the non-neighbors of v from K.
Using these observations they can generate all maximal cliques in G by a recursive algorithm that chooses a vertex v arbitrarily and then, for each maximal clique K in G \ v, outputs both K and the clique formed by adding v to K and removing the non-neighbors of v. However, some cliques of G may be generated in this way from more than one parent clique of G \ v, so they eliminate duplicates by outputting a clique in G only when its parent in G \ v is lexicographically maximum among all possible parent cliques. On the basis of this principle, they show that all maximal cliques in G may be generated in time O(mn) per clique, wherem is the number of edges in G and n is the number of vertices. Chiba & Nishizeki (1985) improve this to O(ma) per clique, where a is the arboricity of the given graph. Makino & Uno (2004) provide an alternative output-sensitive algorithm based on fast matrix multiplication. Johnson & Yannakakis (1988) show that it is even possible to list all maximal cliques in lexicographic order with polynomial delay per clique, although the reverse of this order is NP-hard to generate.
On the basis of this result, it is possible to list all maximal cliques in polynomial time, for families of graphs in which the number of cliques is polynomially bounded. These families include chordal graphs, complete graphs, triangle-free graphs, interval graphs, graphs of bounded boxicity, and planar graphs.[33] In particular, the planar graphs, and more generally, any family of graphs that is both sparse (having a number of edges at most a constant times the number of vertices) and closed under the operation of taking subgraphs, have O(n) cliques, of at most constant size, that can be listed in linear time.[19][34]
Finding maximum cliques in arbitrary graphs
It is possible to find the maximum clique, or the clique number, of an arbitrary n-vertex graph in time O(3n/3) = O(1.4422n) by using one of the algorithms described above to list all maximal cliques in the graph and returning the largest one. However, for this variant of the clique problem better worst-case time bounds are possible. The algorithm of Tarjan & Trojanowski (1977) solves this problem in time O(2n/3) = O(1.2599n). It is a recursive backtracking scheme similar to that of the Bron–Kerbosch algorithm, but is able to eliminate some recursive calls when it can be shown that the cliques found within the call will be suboptimal. Jian (1986) improved the time to O(20.304n) = O(1.2346n), and Robson (1986) improved it to O(20.276n) = O(1.2108n) time, at the expense of greater space usage. Robson's algorithm combines a similar backtracking scheme (with a more complicated case analysis) and a dynamic programming technique in which the optimal solution is precomputed for all small connected subgraphs of the complement graph. These partial solutions are used to shortcut the backtracking recursion. The fastest algorithm known today is a refined version of this method by Robson (2001) which runs in time O(20.249n) = O(1.1888n).[35]
There has also been extensive research on heuristic algorithms for solving maximum clique problems without worst-case runtime guarantees, based on methods including branch and bound,[36] local search,[37] greedy algorithms,[38] and constraint programming.[39] Non-standard computing methodologies for finding cliques include DNA computing[40] and adiabatic quantum computation.[41] The maximum clique problem was the subject of an implementation challenge sponsored by DIMACS in 1992–1993,[42] and a collection of graphs used as benchmarks for the challenge is publicly available.[43]
Special classes of graphs
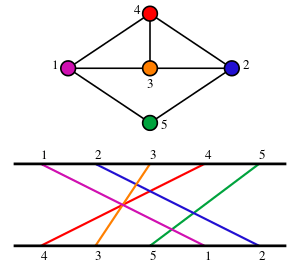
Planar graphs, and other families of sparse graphs, have been discussed above: they have linearly many maximal cliques, of bounded size, that can be listed in linear time.[19] In particular, for planar graphs, any clique can have at most four vertices, by Kuratowski's theorem.[22]
Perfect graphs are defined by the properties that their clique number equals their chromatic number, and that this equality holds also in each of their induced subgraphs. For perfect graphs, it is possible to find a maximum clique in polynomial time, using an algorithm based on semidefinite programming.[44] However, this method is complex and non-combinatorial, and specialized clique-finding algorithms have been developed for many subclasses of perfect graphs.[45] In the complement graphs of bipartite graphs, König's theorem allows the maximum clique problem to be solved using techniques for matching. In another class of perfect graphs, the permutation graphs, a maximum clique is a longest decreasing subsequence of the permutation defining the graph and can be found using known algorithms for the longest decreasing subsequence problem. Conversely, every instance of the longest decreasing subsequence problem can be described equivalently as a problem of finding a maximum clique in a permutation graph.[46] Even, Pnueli & Lempel (1972) provide an alternative quadratic-time algorithm for maximum cliques in comparability graphs, a broader class of perfect graphs that includes the permutation graphs as a special case.[47] In chordal graphs, the maximal cliques can be found by listing the vertices in an elimination ordering, and checking the clique neighborhoods of each vertex in this ordering.[48]
In some cases, these algorithms can be extended to other, non-perfect, classes of graphs as well. For instance, in a circle graph, the neighborhood of each vertex is a permutation graph, so a maximum clique in a circle graph can be found by applying the permutation graph algorithm to each neighborhood.[49] Similarly, in a unit disk graph (with a known geometric representation), there is a polynomial time algorithm for maximum cliques based on applying the algorithm for complements of bipartite graphs to shared neighborhoods of pairs of vertices.[50]
The algorithmic problem of finding a maximum clique in a random graph drawn from the Erdős–Rényi model (in which each edge appears with probability 1/2, independently from the other edges) was suggested by Karp (1976). Because the maximum clique in a random graph has logarithmic size with high probability, it can be found by a brute force search in expected time 2O(log2n). This is a quasi-polynomial time bound.[51] Although the clique number of such graphs is usually very close to 2 log2n, simple greedy algorithms as well as more sophisticated randomized approximation techniques only find cliques with size log2n, half as big. The number of maximal cliques in such graphs is with high probability exponential in log2n, which prevents methods that list all maximal cliques from running in polynomial time.[52] Because of the difficulty of this problem, several authors have investigated the planted clique problem, the clique problem on random graphs that have been augmented by adding large cliques.[53] While spectral methods[54] and semidefinite programming[55] can detect hidden cliques of size Ω(√n), no polynomial-time algorithms are currently known to detect those of size o(√n) (expressed using little-o notation).[56]
Approximation algorithms
Several authors have considered approximation algorithms that attempt to find a clique or independent set that, although not maximum, has size as close to the maximum as can be found in polynomial time. Although much of this work has focused on independent sets in sparse graphs, a case that does not make sense for the complementary clique problem, there has also been work on approximation algorithms that do not use such sparsity assumptions.[57]
Feige (2004) describes a polynomial time algorithm that finds a clique of size Ω((log n/log log n)2) in any graph that has clique number Ω(n/logkn) for any constant k. By using this algorithm when the clique number of a given input graph is between n/log n and n/log3n, switching to a different algorithm of Boppana & Halldórsson (1992) for graphs with higher clique numbers, and choosing a two-vertex clique if both algorithms fail to find anything, Feige provides an approximation algorithm that finds a clique with a number of vertices within a factor of O(n(log log n)2/log3n) of the maximum. Although the approximation ratio of this algorithm is weak, it is the best known to date. The results on hardness of approximation described below suggest that there can be no approximation algorithm with an approximation ratio significantly less than linear.
Lower bounds
NP-completeness
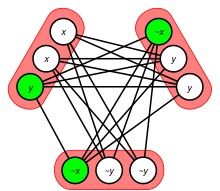
The clique decision problem is NP-complete. It was one of Richard Karp's original 21 problems shown NP-complete in his 1972 paper "Reducibility Among Combinatorial Problems".[59] This problem was also mentioned in Stephen Cook's paper introducing the theory of NP-complete problems.[60] Because of the hardness of the decision problem, the problem of finding a maximum clique is also NP-hard. If one could solve it, one could also solve the decision problem, by comparing the size of the maximum clique to the size parameter given as input in the decision problem.
Karp's NP-completeness proof is a many-one reduction from the Boolean satisfiability problem for formulas in conjunctive normal form, which was proved NP-complete in the Cook–Levin theorem.[61] From a given CNF formula, Karp forms a graph that has a vertex for every pair (v,c), where v is a variable or its negation and c is a clause in the formula that contains v. Two of these vertices are connected by an edge if they represent compatible variable assignments for different clauses. That is, there is an edge from (v,c) to (u,d) whenever c ≠ d and u and v are not each other's negations. If k denotes the number of clauses in the CNF formula, then the k-vertex cliques in this graph represent consistent ways of assigning truth values to some of its variables in order to satisfy the formula. Therefore, the formula is satisfiable if and only if a k-vertex clique exists.[59]
Some NP-complete problems (such as the travelling salesman problem in planar graphs) may be solved in time that is exponential in a sublinear function of the input size parameter n, significantly faster than a brute-force search.[62] However, it is unlikely that such a subexponential time bound is possible for the clique problem in arbitrary graphs, as it would imply similarly subexponential bounds for many other standard NP-complete problems.[63]
Circuit complexity

The computational difficulty of the clique problem has led it to be used to prove several lower bounds in circuit complexity. The existence of a clique of a given size is a monotone graph property, meaning that, if a clique exists in a given graph, it will exist in any supergraph. Because this property is monotone, there must exist a monotone circuit, using only and gates and or gates, to solve the clique decision problem for a given fixed clique size. However, the size of these circuits can be proven to be a super-polynomial function of the number of vertices and the clique size, exponential in the cube root of the number of vertices.[64] Even if a small number of NOT gates are allowed, the complexity remains superpolynomial.[65] Additionally, the depth of a monotone circuit for the clique problem using gates of bounded fan-in must be at least a polynomial in the clique size.[66]
Decision tree complexity
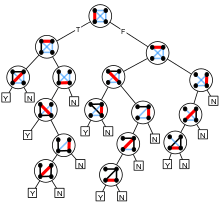
The (deterministic) decision tree complexity of determining a graph property is the number of questions of the form "Is there an edge between vertex u and vertex v?" that have to be answered in the worst case to determine whether a graph has a particular property. That is, it is the minimum height of a boolean decision tree for the problem. There are n(n − 1)/2 possible questions to be asked. Therefore, any graph property can be determined with at most n(n − 1)/2 questions. It is also possible to define random and quantum decision tree complexity of a property, the expected number of questions (for a worst case input) that a randomized or quantum algorithm needs to have answered in order to correctly determine whether the given graph has the property.[67]
Because the property of containing a clique is monotone, it is covered by the Aanderaa–Karp–Rosenberg conjecture, which states that the deterministic decision tree complexity of determining any non-trivial monotone graph property is exactly n(n − 1)/2. For arbitrary monotone graph properties, this conjecture remains unproven. However, for deterministic decision trees, and for any k in the range 2 ≤ k ≤ n, the property of containing a k-clique was shown to have decision tree complexity exactly n(n − 1)/2 by Bollobás (1976). Deterministic decision trees also require exponential size to detect cliques, or large polynomial size to detect cliques of bounded size.[68]
The Aanderaa–Karp–Rosenberg conjecture also states that the randomized decision tree complexity of non-trivial monotone functions is Θ(n2). The conjecture again remains unproven, but has been resolved for the property of containing a k clique for 2 ≤ k ≤ n. This property is known to have randomized decision tree complexity Θ(n2).[69] For quantum decision trees, the best known lower bound is Ω(n), but no matching algorithm is known for the case of k ≥ 3.[70]
Fixed-parameter intractability
Parameterized complexity is the complexity-theoretic study of problems that are naturally equipped with a small integer parameter k and for which the problem becomes more difficult as k increases, such as finding k-cliques in graphs. A problem is said to be fixed-parameter tractable if there is an algorithm for solving it on inputs of size n, and a function f, such that the algorithm runs in time f(k) nO(1). That is, it is fixed-parameter tractable if it can be solved in polynomial time for any fixed value of k and moreover if the exponent of the polynomial does not depend on k.[71]
For finding k-vertex cliques, the brute force search algorithm has running time O(nkk2). Because the exponent of n depends on k, this algorithm is not fixed-parameter tractable. Although it can be improved by fast matrix multiplication the running time still has an exponent that is linear in k Thus, although the running time of known algorithms for the clique problem is polynomial for any fixed k these algorithms do not suffice for fixed-parameter tractability. Downey & Fellows (1995) defined a hierarchy of parametrized problems, the W hierarchy, that they conjectured did not have fixed-parameter tractable algorithms. They proved that independent set (or, equivalently, clique) is hard for the first level of this hierarchy, W[1]. Thus, according to their conjecture, clique has no fixed-parameter tractable algorithm. Moreover, this result provides the basis for proofs of W[1]-hardness of many other problems, and thus serves as an analogue of the Cook–Levin theorem for parameterized complexity.[72]
Chen et al. (2006) showed that finding k-vertex cliques cannot be done in time no(k) unless the exponential time hypothesis fails. Again, this provides evidence that no fixed-parameter tractable algorithm is possible.[73]
Although the problems of listing maximal cliques or finding maximum cliques are unlikely to be fixed-parameter tractable with the parameter k, they may be fixed-parameter tractable for other parameters of instance complexity. For instance, both problems are known to be fixed-parameter tractable when parametrized by the degeneracy of the input graph.[34]
Hardness of approximation
Weak results hinting that the clique problem might be hard to approximate have been known for a long time. Garey & Johnson (1978) observed that, because of the fact that the clique number takes on small integer values and is NP-hard to compute, it cannot have a fully polynomial-time approximation scheme. If too accurate an approximation were available, rounding its value to an integer would give the exact clique number. However, little more was known until the early 1990s, when several authors began to make connections between the approximation of maximum cliques and probabilistically checkable proofs. They used these connections to prove hardness of approximation results for the maximum clique problem.[74] After many improvements to these results it is now known that, unless P = NP, there can be no polynomial time algorithm that approximates the maximum clique to within a factor better than O(n1 − ε), for any ε > 0.[75]
The rough idea of these inapproximability results is to form a graph that represents a probabilistically checkable proof system for an NP-complete problem such as the Boolean satisfiability problem. In a probabilistically checkable proof system, a proof is represented as a sequence of bits. It is checked by an algorithm that, after a polynomial-time computation on the input to the satisfiability problem, chooses to examine a small number of randomly chosen positions of the proof string. Depending on what values are found at that sample of bits, the checker will either accept or reject the proof, without looking at the rest of the bits. False negatives are not allowed: a valid proof must always be accepted. However, an invalid proof may sometimes mistakenly be accepted, as long as the probability that a checker makes a mistake of this type is low. An instance of the satisfiability problem should have a valid proof if and only if it is satisfiable.[76]
To transform a probabilistically checkable proof system of this type into a clique problem, one forms a graph with a vertex for each possible accepting run of the proof checker. That is, a vertex is defined by one of the possible random choices of sets of positions to examine, and by bit values for those positions that would cause the checker to accept the proof. Two vertices are adjacent, in this graph, if they agree on the value of each bit at the positions they have in common. Because of this construction, the maximal cliques in this graph correspond to possible proof strings. The clique for a proof string is the set of accepting runs of a checker on that proof string, regardless of whether it actually is a valid proof or not. One of these cliques is large if and only if it corresponds to a proof string that many proof checkers accept. If the original satisfiability instance is satisfiable, it will have a valid proof string, one that is accepted by all runs of the checker, and this string will correspond to a large clique in the graph. However, if the original instance is not satisfiable, then all proof strings are invalid, each proof string has only a small number of checker runs that mistakenly accept it, and all cliques are small. Therefore, if one could distinguish in polynomial time between graphs that have large cliques and graphs in which all cliques are small, or if one could accurately approximate the clique problem, then applying this approximation to the graphs generated from satisfiability instances would allow satisfiable instances to be distinguished from unsatisfiable instances. However, this is not possible unless P = NP.[76]
Notes
- 1 2 3 Bomze et al. (1999); Gutin (2004).
- ↑ Wasserman & Faust (1994).
- ↑ Kolata (1990).
- ↑ Rhodes et al. (2003).
- ↑ Kuhl, Crippen & Friesen (1983).
- ↑ National Research Council Committee on Mathematical Challenges from Computational Chemistry (1995). See in particular pp. 35–36.
- ↑ Muegge & Rarey (2001). See in particular pp. 6–7.
- ↑ Barrow & Burstall (1976).
- ↑ Hamzaoglu & Patel (1998).
- ↑ Day & Sankoff (1986).
- ↑ Samudrala & Moult (1998).
- ↑ Spirin & Mirny (2003).
- ↑ Frank & Strauss (1986).
- ↑ The Keller graph used by Lagarias & Shor (1992) has 1048576 vertices and clique size 1024. They described a synthetic construction for the clique, but also used clique-finding algorithms on smaller graphs to help guide their search. Mackey (2002) simplified the proof by finding a clique of size 256 in a 65536-vertex Keller graph.
- 1 2 Valiente (2002); Pelillo (2009).
- ↑ Pelillo (2009).
- ↑ Sethuraman & Butenko (2015).
- ↑ & Valiente (2002).
- 1 2 3 4 Chiba & Nishizeki (1985).
- 1 2 Cormen et al. (2001).
- ↑ Cormen et al. (2001), Exercise 34-1, p. 1018.
- 1 2 Papadimitriou & Yannakakis (1981); Chiba & Nishizeki (1985).
- ↑ Garey, Johnson & Stockmeyer (1976).
- ↑ See, e.g., Frank & Strauss (1986).
- ↑ Plummer (1993).
- ↑ Skiena (2009), p. 526.
- ↑ Cook (1985).
- ↑ E.g., see Downey & Fellows (1995).
- ↑ Itai & Rodeh (1978) provide an algorithm with O(m3/2) running time that finds a triangle if one exists but does not list all triangles; Chiba & Nishizeki (1985) list all triangles in time O(m3/2).
- ↑ Eisenbrand & Grandoni (2004); Kloks, Kratsch & Müller (2000); Nešetřil & Poljak (1985); Vassilevska & Williams (2009); Yuster (2006).
- ↑ Tomita, Tanaka & Takahashi (2006).
- ↑ Cazals & Karande (2008); Eppstein, Löffler & Strash (2013).
- ↑ Rosgen & Stewart (2007).
- 1 2 Eppstein, Löffler & Strash (2013).
- ↑ Robson (2001).
- ↑ Balas & Yu (1986); Carraghan & Pardalos (1990); Pardalos & Rogers (1992); Östergård (2002); Fahle (2002); Tomita & Seki (2003); Tomita & Kameda (2007); Konc & Janežič (2007).
- ↑ Battiti & Protasi (2001); Katayama, Hamamoto & Narihisa (2005).
- ↑ Abello, Pardalos & Resende (1999); Grosso, Locatelli & Della Croce (2004).
- ↑ Régin (2003).
- ↑ Ouyang et al. (1997). Although the title refers to maximal cliques, the problem this paper solves is actually the maximum clique problem.
- ↑ Childs et al. (2002).
- ↑ Johnson & Trick (1996).
- ↑ DIMACS challenge graphs for the clique problem, accessed 2009-12-17.
- ↑ Grötschel, Lovász & Schrijver (1988).
- ↑ Golumbic (1980).
- ↑ Golumbic (1980), p. 159.
- ↑ Even, Pnueli & Lempel (1972).
- ↑ Blair & Peyton (1993), Lemma 4.5, p. 19.
- ↑ Gavril (1973); Golumbic (1980), p. 247.
- ↑ Clark, Colbourn & Johnson (1990).
- ↑ Song (2015).
- ↑ Jerrum (1992).
- ↑ Arora & Barak (2009), Example 18.2, pp. 362–363.
- ↑ Alon, Krivelevich & Sudakov (1998).
- ↑ Feige & Krauthgamer (2000).
- ↑ Meka, Potechin & Wigderson (2015).
- ↑ Boppana & Halldórsson (1992); Feige (2004); Halldórsson (2000).
- ↑ Adapted from Sipser (1996)
- 1 2 Karp (1972).
- ↑ Cook (1971).
- ↑ Cook (1971) gives essentially the same reduction, from 3-SAT instead of Satisfiability, to show that subgraph isomorphism is NP-complete.
- ↑ Lipton & Tarjan (1980).
- ↑ Impagliazzo, Paturi & Zane (2001).
- ↑ Alon & Boppana (1987). For earlier and weaker bounds on monotone circuits for the clique problem, see Valiant (1983) and Razborov (1985).
- ↑ Amano & Maruoka (1998).
- ↑ Goldmann & Håstad (1992) used communication complexity to prove this result.
- ↑ See Arora & Barak (2009), Chapter 12, "Decision trees", pp. 259–269.
- ↑ Wegener (1988).
- ↑ For instance, this follows from Gröger (1992).
- ↑ Childs & Eisenberg (2005); Magniez, Santha & Szegedy (2007).
- ↑ Downey & Fellows (1999). Technically, there is usually an additional requirement that f be a computable function.
- ↑ Downey & Fellows (1995).
- ↑ Chen et al. (2006).
- ↑ Kolata (1990); Feige et al. (1991); Arora & Safra (1998); Arora et al. (1998).
- ↑ Håstad (1999) showed inapproximability for this ratio using a stronger complexity theoretic assumption, the inequality of NP and ZPP. Khot (2001) described more precisely the inapproximability ratio, but with an even stronger assumption. Zuckerman (2006) derandomized the construction weakening its assumption to P ≠ NP.
- 1 2 This reduction is originally due to Feige et al. (1991) and used in all subsequent inapproximability proofs; the proofs differ in the strengths and details of the probabilistically checkable proof systems that they rely on.
References
Surveys and textbooks
- Arora, Sanjeev; Barak, Boaz (2009), Computational Complexity: A Modern Approach, Cambridge University Press, ISBN 978-0-521-42426-4.
- Blair, Jean R. S.; Peyton, Barry (1993), "An introduction to chordal graphs and clique trees", Graph theory and sparse matrix computation, IMA Vol. Math. Appl., 56, Springer, New York, pp. 1–29, doi:10.1007/978-1-4613-8369-7_1, MR 1320296.
- Bomze, I. M.; Budinich, M.; Pardalos, P. M.; Pelillo, M. (1999), "The maximum clique problem", Handbook of Combinatorial Optimization, 4, Kluwer Academic Publishers, pp. 1–74, CiteSeerX 10.1.1.48.4074
 .
. - Cormen, Thomas H.; Leiserson, Charles E.; Rivest, Ronald L.; Stein, Clifford (2001), "34.5.1 The clique problem", Introduction to Algorithms (2nd ed.), MIT Press and McGraw-Hill, pp. 1003–1006, ISBN 0-262-03293-7.
- Downey, R. G.; Fellows, M. R. (1999), Parameterized complexity, Springer-Verlag, ISBN 0-387-94883-X.
- Golumbic, M. C. (1980), Algorithmic Graph Theory and Perfect Graphs, Computer Science and Applied Mathematics, Academic Press, ISBN 0-444-51530-5.
- Grötschel, M.; Lovász, L.; Schrijver, A. (1988), "9.4 Coloring Perfect Graphs", Geometric Algorithms and Combinatorial Optimization, Algorithms and Combinatorics, 2, Springer-Verlag, pp. 296–298, ISBN 0-387-13624-X.
- Gutin, G. (2004), "5.3 Independent sets and cliques", in Gross, J. L.; Yellen, J., Handbook of graph theory, Discrete Mathematics & Its Applications, CRC Press, pp. 389–402, ISBN 978-1-58488-090-5.
- Muegge, Ingo; Rarey, Matthias (2001), "Small molecule docking and scoring", Reviews in Computational Chemistry, 17: 1–60, doi:10.1002/0471224413.ch1.
- National Research Council Committee on Mathematical Challenges from Computational Chemistry (1995), Mathematical Challenges from Theoretical/Computational Chemistry, National Academies Press, doi:10.17226/4886.
- Pelillo, Marcello (2009), "Heuristics for maximum clique and independent set", Encyclopedia of Optimization, Springer, pp. 1508–1520, doi:10.1007/978-0-387-74759-0_264.
- Plummer, Michael D. (1993), "Well-covered graphs: a survey", Quaestiones Mathematicae, 16 (3): 253–287, doi:10.1080/16073606.1993.9631737, MR 1254158.
- Sipser, M. (1996), Introduction to the Theory of Computation, International Thompson Publishing, ISBN 0-534-94728-X.
- Skiena, Steven S. (2009), The Algorithm Design Manual (2nd ed.), Springer, ISBN 978-1-84800-070-4.
- Valiente, Gabriel (2002), "Chapter 6: Clique, Independent Set, and Vertex Cover", Algorithms on Trees and Graphs, Springer, pp. 299–350, doi:10.1007/978-3-662-04921-1_6.
- Wasserman, Stanley; Faust, Katherine (1994), Social Network Analysis: Methods and Applications, Structural Analysis in the Social Sciences, 8, Cambridge University Press, p. 276, ISBN 978-0-521-38707-1.
Popular press
- Kolata, Gina (June 26, 1990), "In a Frenzy, Math Enters Age of Electronic Mail", New York Times.
Research articles
- Abello, J.; Pardalos, P. M.; Resende, M. G. C. (1999), "On maximum clique problems in very large graphs", in Abello, J.; Vitter, J., External Memory Algorithms, DIMACS Series on Discrete Mathematics and Theoretical Computer Science, 50, American Mathematical Society, pp. 119–130, ISBN 0-8218-1184-3.
- Alon, N.; Boppana, R. (1987), "The monotone circuit complexity of boolean functions", Combinatorica, 7 (1): 1–22, doi:10.1007/BF02579196.
- Alon, N.; Krivelevich, M.; Sudakov, B. (1998), "Finding a large hidden clique in a random graph", Random Structures & Algorithms, 13 (3–4): 457–466, doi:10.1002/(SICI)1098-2418(199810/12)13:3/4<457::AID-RSA14>3.0.CO;2-W.
- Alon, N.; Yuster, R.; Zwick, U. (1994), "Finding and counting given length cycles", Proceedings of the 2nd European Symposium on Algorithms, Utrecht, The Netherlands, pp. 354–364.
- Amano, K.; Maruoka, A. (1998), "A superpolynomial lower bound for a circuit computing the clique function with at most (1/6)log log n negation gates", Proc. Symp. Mathematical Foundations of Computer Science, Lecture Notes in Computer Science, 1450, Springer-Verlag, pp. 399–408.
- Arora, Sanjeev; Lund, Carsten; Motwani, Rajeev; Sudan, Madhu; Szegedy, Mario (1998), "Proof verification and the hardness of approximation problems", Journal of the ACM, 45 (3): 501–555, doi:10.1145/278298.278306, ECCC TR98-008. Originally presented at the 1992 Symposium on Foundations of Computer Science, doi:10.1109/SFCS.1992.267823.
- Arora, S.; Safra, S. (1998), "Probabilistic checking of proofs: A new characterization of NP", Journal of the ACM, 45 (1): 70–122, doi:10.1145/273865.273901. Originally presented at the 1992 Symposium on Foundations of Computer Science, doi:10.1109/SFCS.1992.267824.
- Balas, E.; Yu, C. S. (1986), "Finding a maximum clique in an arbitrary graph", SIAM Journal on Computing, 15 (4): 1054–1068, doi:10.1137/0215075.
- Barrow, H.; Burstall, R. (1976), "Subgraph isomorphism, matching relational structures and maximal cliques", Information Processing Letters, 4 (4): 83–84, doi:10.1016/0020-0190(76)90049-1.
- Battiti, R.; Protasi, M. (2001), "Reactive local search for the maximum clique problem", Algorithmica, 29 (4): 610–637, doi:10.1007/s004530010074.
- Bollobás, Béla (1976), "Complete subgraphs are elusive", Journal of Combinatorial Theory, Series B, 21 (1): 1–7, doi:10.1016/0095-8956(76)90021-6, ISSN 0095-8956.
- Boppana, R.; Halldórsson, M. M. (1992), "Approximating maximum independent sets by excluding subgraphs", BIT Numerical Mathematics, 32 (2): 180–196, doi:10.1007/BF01994876.
- Bron, C.; Kerbosch, J. (1973), "Algorithm 457: finding all cliques of an undirected graph", Communications of the ACM, 16 (9): 575–577, doi:10.1145/362342.362367.
- Carraghan, R.; Pardalos, P. M. (1990), "An exact algorithm for the maximum clique problem" (PDF), Operations Research Letters, 9 (6): 375–382, doi:10.1016/0167-6377(90)90057-C.
- Cazals, F.; Karande, C. (2008), "A note on the problem of reporting maximal cliques" (PDF), Theoretical Computer Science, 407 (1): 564–568, doi:10.1016/j.tcs.2008.05.010.
- Chen, Jianer; Huang, Xiuzhen; Kanj, Iyad A.; Xia, Ge (2006), "Strong computational lower bounds via parameterized complexity", Journal of Computer and System Sciences, 72 (8): 1346–1367, doi:10.1016/j.jcss.2006.04.007
- Chiba, N.; Nishizeki, T. (1985), "Arboricity and subgraph listing algorithms", SIAM Journal on Computing, 14 (1): 210–223, doi:10.1137/0214017.
- Childs, A. M.; Farhi, E.; Goldstone, J.; Gutmann, S. (2002), "Finding cliques by quantum adiabatic evolution", Quantum Information and Computation, 2 (3): 181–191, arXiv:quant-ph/0012104
 .
. - Childs, A. M.; Eisenberg, J. M. (2005), "Quantum algorithms for subset finding", Quantum Information and Computation, 5 (7): 593–604, arXiv:quant-ph/0311038
 .
. - Clark, Brent N.; Colbourn, Charles J.; Johnson, David S. (1990), "Unit disk graphs", Discrete Mathematics, 86 (1–3): 165–177, doi:10.1016/0012-365X(90)90358-O
- Cook, S. A. (1971), "The complexity of theorem-proving procedures", Proc. 3rd ACM Symposium on Theory of Computing, pp. 151–158, doi:10.1145/800157.805047.
- Cook, Stephen A. (1985), "A taxonomy of problems with fast parallel algorithms", Information and Control, 64 (1–3): 2–22, doi:10.1016/S0019-9958(85)80041-3, MR 837088.
- Day, William H. E.; Sankoff, David (1986), "Computational complexity of inferring phylogenies by compatibility", Systematic Zoology, 35 (2): 224–229, doi:10.2307/2413432, JSTOR 2413432.
- Downey, R. G.; Fellows, M. R. (1995), "Fixed-parameter tractability and completeness. II. On completeness for W[1]", Theoretical Computer Science, 141 (1–2): 109–131, doi:10.1016/0304-3975(94)00097-3.
- Eisenbrand, F.; Grandoni, F. (2004), "On the complexity of fixed parameter clique and dominating set", Theoretical Computer Science, 326 (1–3): 57–67, doi:10.1016/j.tcs.2004.05.009.
- Eppstein, David; Löffler, Maarten; Strash, Darren (2013), "Listing all maximal cliques in large sparse real-world graphs in near-optimal time", Journal of Experimental Algorithmics, 18 (3): 3.1, doi:10.1145/2543629.
- Erdős, Paul; Szekeres, George (1935), "A combinatorial problem in geometry" (PDF), Compositio Mathematica, 2: 463–470.
- Even, S.; Pnueli, A.; Lempel, A. (1972), "Permutation graphs and transitive graphs", Journal of the ACM, 19 (3): 400–410, doi:10.1145/321707.321710.
- Fahle, T. (2002), "Simple and fast: Improving a branch-and-bound algorithm for maximum clique", Proc. 10th European Symposium on Algorithms, Lecture Notes in Computer Science, 2461, Springer-Verlag, pp. 47–86, doi:10.1007/3-540-45749-6_44, ISBN 978-3-540-44180-9.
- Feige, U. (2004), "Approximating maximum clique by removing subgraphs", SIAM Journal on Discrete Mathematics, 18 (2): 219–225, doi:10.1137/S089548010240415X.
- Feige, U.; Goldwasser, S.; Lovász, L.; Safra, S; Szegedy, M. (1991), "Approximating clique is almost NP-complete", Proc. 32nd IEEE Symp. on Foundations of Computer Science, pp. 2–12, doi:10.1109/SFCS.1991.185341, ISBN 0-8186-2445-0.
- Feige, U.; Krauthgamer, R. (2000), "Finding and certifying a large hidden clique in a semirandom graph", Random Structures and Algorithms, 16 (2): 195–208, doi:10.1002/(SICI)1098-2418(200003)16:2<195::AID-RSA5>3.0.CO;2-A.
- Frank, Ove; Strauss, David (1986), "Markov graphs", Journal of the American Statistical Association, 81 (395): 832–842, JSTOR 2289017, MR 860518.
- Garey, M. R.; Johnson, D. S. (1978), ""Strong" NP-completeness results: motivation, examples and implications", Journal of the ACM, 25 (3): 499–508, doi:10.1145/322077.322090.
- Garey, M. R.; Johnson, D. S.; Stockmeyer, L. (1976), "Some simplified NP-complete graph problems", Theoretical Computer Science, 1 (3): 237–267, doi:10.1016/0304-3975(76)90059-1, MR 0411240.
- Gavril, F. (1973), "Algorithms for a maximum clique and a maximum independent set of a circle graph", Networks, 3 (3): 261–273, doi:10.1002/net.3230030305.
- Goldmann, M.; Håstad, J. (1992), "A simple lower bound for monotone clique using a communication game", Information Processing Letters, 41 (4): 221–226, doi:10.1016/0020-0190(92)90184-W.
- Gröger, Hans Dietmar (1992), "On the randomized complexity of monotone graph properties" (PDF), Acta Cybernetica, 10 (3): 119–127, retrieved 2009-10-02
- Grosso, A.; Locatelli, M.; Della Croce, F. (2004), "Combining swaps and node weights in an adaptive greedy approach for the maximum clique problem", Journal of Heuristics, 10 (2): 135–152, doi:10.1023/B:HEUR.0000026264.51747.7f.
- Halldórsson, M. M. (2000), "Approximations of Weighted Independent Set and Hereditary Subset Problems" (PDF), Journal of Graph Algorithms and Applications, 4 (1): 1–16, doi:10.7155/jgaa.00020.
- Hamzaoglu, I.; Patel, J. H. (1998), "Test set compaction algorithms for combinational circuits", Proc. 1998 IEEE/ACM International Conference on Computer-Aided Design, pp. 283–289, doi:10.1145/288548.288615.
- Harary, F.; Ross, I. C. (1957), "A procedure for clique detection using the group matrix", Sociometry, American Sociological Association, 20 (3): 205–215, doi:10.2307/2785673, JSTOR 2785673, MR 0110590.
- Håstad, J. (1999), "Clique is hard to approximate within n1 − ε", Acta Mathematica, 182 (1): 105–142, doi:10.1007/BF02392825.
- Impagliazzo, R.; Paturi, R.; Zane, F. (2001), "Which problems have strongly exponential complexity?", Journal of Computer and System Sciences, 63 (4): 512–530, doi:10.1006/jcss.2001.1774.
- Itai, A.; Rodeh, M. (1978), "Finding a minimum circuit in a graph", SIAM Journal on Computing, 7 (4): 413–423, doi:10.1137/0207033.
- Jerrum, M. (1992), "Large cliques elude the Metropolis process", Random Structures and Algorithms, 3 (4): 347–359, doi:10.1002/rsa.3240030402.
- Jian, T (1986), "An O(20.304n) algorithm for solving maximum independent set problem", IEEE Transactions on Computers, IEEE Computer Society, 35 (9): 847–851, doi:10.1109/TC.1986.1676847, ISSN 0018-9340.
- Johnson, D. S.; Trick, M. A., eds. (1996), Cliques, Coloring, and Satisfiability: Second DIMACS Implementation Challenge, October 11–13, 1993, DIMACS Series in Discrete Mathematics and Theoretical Computer Science, 26, American Mathematical Society, ISBN 0-8218-6609-5.
- Johnson, D. S.; Yannakakis, M. (1988), "On generating all maximal independent sets", Information Processing Letters, 27 (3): 119–123, doi:10.1016/0020-0190(88)90065-8.
- Karp, Richard M. (1972), "Reducibility among combinatorial problems", in Miller, R. E.; Thatcher, J. W., Complexity of Computer Computations (PDF), New York: Plenum, pp. 85–103.
- Karp, Richard M. (1976), "Probabilistic analysis of some combinatorial search problems", in Traub, J. F., Algorithms and Complexity: New Directions and Recent Results, New York: Academic Press, pp. 1–19.
- Katayama, K.; Hamamoto, A.; Narihisa, H. (2005), "An effective local search for the maximum clique problem", Information Processing Letters, 95 (5): 503–511, doi:10.1016/j.ipl.2005.05.010.
- Khot, S. (2001), "Improved inapproximability results for MaxClique, chromatic number and approximate graph coloring", Proc. 42nd IEEE Symp. Foundations of Computer Science, pp. 600–609, doi:10.1109/SFCS.2001.959936, ISBN 0-7695-1116-3.
- Kloks, T.; Kratsch, D.; Müller, H. (2000), "Finding and counting small induced subgraphs efficiently", Information Processing Letters, 74 (3–4): 115–121, doi:10.1016/S0020-0190(00)00047-8.
- Konc, J.; Janežič, D. (2007), "An improved branch and bound algorithm for the maximum clique problem" (PDF), MATCH Communications in Mathematical and in Computer Chemistry, 58 (3): 569–590. Source code
- Kuhl, F. S.; Crippen, G. M.; Friesen, D. K. (1983), "A combinatorial algorithm for calculating ligand binding", Journal of Computational Chemistry, 5 (1): 24–34, doi:10.1002/jcc.540050105.
- Lagarias, Jeffrey C.; Shor, Peter W. (1992), "Keller's cube-tiling conjecture is false in high dimensions", Bulletin of the American Mathematical Society, New Series, 27 (2): 279–283, doi:10.1090/S0273-0979-1992-00318-X, MR 1155280.
- Lipton, R. J.; Tarjan, R. E. (1980), "Applications of a planar separator theorem", SIAM Journal on Computing, 9 (3): 615–627, doi:10.1137/0209046.
- Luce, R. Duncan; Perry, Albert D. (1949), "A method of matrix analysis of group structure", Psychometrika, 14 (2): 95–116, doi:10.1007/BF02289146, PMID 18152948.
- Mackey, John (2002), "A cube tiling of dimension eight with no facesharing", Discrete and Computational Geometry, 28 (2): 275–279, doi:10.1007/s00454-002-2801-9, MR 1920144.
- Magniez, Frédéric; Santha, Miklos; Szegedy, Mario (2007), "Quantum algorithms for the triangle problem", SIAM Journal on Computing, 37 (2): 413–424, arXiv:quant-ph/0310134
 , doi:10.1137/050643684.
, doi:10.1137/050643684. - Makino, K.; Uno, T. (2004), "New algorithms for enumerating all maximal cliques", Algorithm Theory: SWAT 2004, Lecture Notes in Computer Science, 3111, Springer-Verlag, pp. 260–272.
- Meka, Raghu; Potechin, Aaron; Wigderson, Avi (2015), "Sum-of-squares lower bounds for planted clique", Proceedings of the Forty-Seventh Annual ACM on Symposium on Theory of Computing (STOC '15), New York, NY, USA: ACM, pp. 87–96, arXiv:1503.06447
 , doi:10.1145/2746539.2746600, ISBN 978-1-4503-3536-2.
, doi:10.1145/2746539.2746600, ISBN 978-1-4503-3536-2. - Moon, J. W.; Moser, L. (1965), "On cliques in graphs", Israel Journal of Mathematics, 3: 23–28, doi:10.1007/BF02760024, MR 0182577.
- Nešetřil, J.; Poljak, S. (1985), "On the complexity of the subgraph problem", Commentationes Mathematicae Universitatis Carolinae, 26 (2): 415–419.
- Östergård, P. R. J. (2002), "A fast algorithm for the maximum clique problem", Discrete Applied Mathematics, 120 (1–3): 197–207, doi:10.1016/S0166-218X(01)00290-6.
- Ouyang, Q.; Kaplan, P. D.; Liu, S.; Libchaber, A. (1997), "DNA solution of the maximal clique problem", Science, 278 (5337): 446–449, doi:10.1126/science.278.5337.446, PMID 9334300.
- Papadimitriou, Christos H.; Yannakakis, Mihalis (1981), "The clique problem for planar graphs", Information Processing Letters, 13 (4–5): 131–133, doi:10.1016/0020-0190(81)90041-7, MR 651460.
- Pardalos, P. M.; Rogers, G. P. (1992), "A branch and bound algorithm for the maximum clique problem", Computers & Operations Research, 19 (5): 363–375, doi:10.1016/0305-0548(92)90067-F.
- Razborov, A. A. (1985), "Lower bounds for the monotone complexity of some Boolean functions", Proceedings of the USSR Academy of Sciences (in Russian), 281: 798–801. English translation in Sov. Math. Dokl. 31 (1985): 354–357.
- Régin, J.-C. (2003), "Using constraint programming to solve the maximum clique problem", Proc. 9th Int. Conf. Principles and Practice of Constraint Programming – CP 2003, Lecture Notes in Computer Science, 2833, Springer-Verlag, pp. 634–648.
- Rhodes, Nicholas; Willett, Peter; Calvet, Alain; Dunbar, James B.; Humblet, Christine (2003), "CLIP: similarity searching of 3D databases using clique detection", Journal of Chemical Information and Computer Sciences, 43 (2): 443–448, doi:10.1021/ci025605o, PMID 12653507.
- Robson, J. M. (1986), "Algorithms for maximum independent sets", Journal of Algorithms, 7 (3): 425–440, doi:10.1016/0196-6774(86)90032-5.
- Robson, J. M. (2001), Finding a maximum independent set in time O(2n/4).
- Rosgen, B; Stewart, L (2007), "Complexity results on graphs with few cliques", Discrete Mathematics and Theoretical Computer Science, 9 (1): 127–136.
- Samudrala, Ram; Moult, John (1998), "A graph-theoretic algorithm for comparative modeling of protein structure", Journal of Molecular Biology, 279 (1): 287–302, doi:10.1006/jmbi.1998.1689, PMID 9636717.
- Sethuraman, Samyukta; Butenko, Sergiy (2015), "The maximum ratio clique problem", Computational Management Science, 12 (1): 197–218, doi:10.1007/s10287-013-0197-z, MR 3296231.
- Song, Y. (2015), "On the independent set problem in random graphs", International Journal of Computer Mathematics, 92 (11): 2233–2242.
- Spirin, Victor; Mirny, Leonid A. (2003), "Protein complexes and functional modules in molecular networks", Proceedings of the National Academy of Sciences, 100 (21): 12123–12128, doi:10.1073/pnas.2032324100, PMC 218723
 , PMID 14517352.
, PMID 14517352. - Tarjan, R. E.; Trojanowski, A. E. (1977), "Finding a maximum independent set" (PDF), SIAM Journal on Computing, 6 (3): 537–546, doi:10.1137/0206038.
- Tomita, E.; Kameda, T. (2007), "An efficient branch-and-bound algorithm for finding a maximum clique with computational experiments", Journal of Global Optimization, 37 (1): 95–111, doi:10.1007/s10898-006-9039-7.
- Tomita, E.; Seki, T. (2003), "An efficient branch-and-bound algorithm for finding a maximum clique", Discrete Mathematics and Theoretical Computer Science, Lecture Notes in Computer Science, 2731, Springer-Verlag, pp. 278–289, doi:10.1007/3-540-45066-1_22, ISBN 978-3-540-40505-4.
- Tomita, E.; Tanaka, A.; Takahashi, H. (2006), "The worst-case time complexity for generating all maximal cliques and computational experiments", Theoretical Computer Science, 363 (1): 28–42, doi:10.1016/j.tcs.2006.06.015.
- Tsukiyama, S.; Ide, M.; Ariyoshi, I.; Shirakawa, I. (1977), "A new algorithm for generating all the maximal independent sets", SIAM Journal on Computing, 6 (3): 505–517, doi:10.1137/0206036.
- Valiant, L. G. (1983), "Exponential lower bounds for restricted monotone circuits", Proc. 15th ACM Symposium on Theory of Computing, pp. 110–117, doi:10.1145/800061.808739, ISBN 0-89791-099-0.
- Vassilevska, V.; Williams, R. (2009), "Finding, minimizing, and counting weighted subgraphs", Proc. 41st ACM Symposium on Theory of Computing, pp. 455–464, doi:10.1145/1536414.1536477, ISBN 978-1-60558-506-2.
- Wegener, I. (1988), "On the complexity of branching programs and decision trees for clique functions", Journal of the ACM, 35 (2): 461–472, doi:10.1145/42282.46161.
- Yuster, R. (2006), "Finding and counting cliques and independent sets in r-uniform hypergraphs", Information Processing Letters, 99 (4): 130–134, doi:10.1016/j.ipl.2006.04.005.
- Zuckerman, D. (2006), "Linear degree extractors and the inapproximability of max clique and chromatic number", Proc. 38th ACM Symp. Theory of Computing, pp. 681–690, doi:10.1145/1132516.1132612, ISBN 1-59593-134-1, ECCC TR05-100.