Homologous somatic pairing
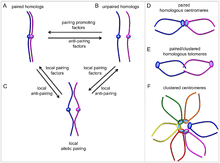
Somatic pairing of homologous chromosomes is similar to pre- and early meiotic pairing (see article: Homologous chromosome#In meiosis), and has been observed in Diptera[1] (Drosophila), and budding yeast,[2] for example (whether it evolved multiple times in metazoans is unclear[3]). Mammals show little pairing apart from in germline cells, taking place at specific loci, and under the control of developmental signalling (understood as a subset of other long-range interchromosomal interactions such as looping, and organisation into chromosomal territories).[4]
While meiotic pairing has been extensively studied, the role of somatic pairing has remained less well understood, and even whether it is mechanistically related to meiotic pairing is unknown.[5]
Early work
The first review of somatic pairing was made by Metz in 1916,[1] citing the first descriptions of pairing made in 1907[6] and 1908 by N. M. Stevens in germline cells, who noted:[7]
“it may therefore be true that pairing of homologous chromosomes occurs in connection with each mitosis throughout the life history of these insects” (p.215)
Stevens noted the potential for communication and a role in heredity.[7]
While meiotic homologous pairing subsequently became well studied, somatic pairing remained neglected due to what has been described as "limitations in cytological tools for measuring pairing and genetic tools for perturbing pairing dynamics".[5]
Recent findings from high-throughput screening
In 1998 it was determined that homologous pairing in Drosophila occurs through independent initiations (as opposed to a directed, 'processive zippering' motion).[4][8]
The first RNAi screen (based on DNA FISH[9]) was carried out to identify genes regulating D. melanogaster somatic pairing in 2012,[10] described at the time as providing "an extensive “parts list” of mostly novel factors". These comprised 40 pairing promoting genes and 65 'anti-pairing' genes (of which 2 and 1 were already known, respectively), many of which have human orthologs.[5]
An earlier RNAi screen in 2007 showed the disruption of Topoisomerase II activity impairs somatic pairing within Drosophila tissue culture,[11] indicating a role for topoisomerase-mediated organisation (or the direct interactions of topoisomerase enzymes) in pairing.[4] Condensin (despite dependent interactions with Topoisomerase II) is antagonistic to Drosophila homologous pairing.[12]
References
- 1 2 Metz, Charles W. (1916). "Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance". Journal of Experimental Zoology. 21 (2): 213–279. doi:10.1002/jez.1400210204. ISSN 0022-104X.
- ↑ Burgess, Sean M; Kleckner, Nancy; Weiner, Beth M (1999). "Somatic pairing of homologs in budding yeast: existence and modulation". Genes & Development. 13: 1627–1641. doi:10.1101/gad.13.12.1627. ISSN 1549-5477.
- ↑ Joyce, Eric F; Erceg, Jelena; Wu, C-ting (2016). "Pairing and anti-pairing: a balancing act in the diploid genome". Current Opinion in Genetics & Development. 37: 119–128. doi:10.1016/j.gde.2016.03.002. ISSN 0959-437X.
- 1 2 3 Apte, Manasi S.; Meller, Victoria H. (2012). "Homologue Pairing in Flies and Mammals: Gene Regulation When Two Are Involved". Genetics Research International. 2012: 1–9. doi:10.1155/2012/430587. ISSN 2090-3154.
- 1 2 3 Copenhaver, Gregory P.; Bosco, Giovanni (2012). "Chromosome Pairing: A Hidden Treasure No More". PLoS Genetics. 8 (5): e1002737. doi:10.1371/journal.pgen.1002737. ISSN 1553-7404.
- ↑ Stevens, N. M. (1907) The chromosomes of Drosophila ampelophila. Proc. VII Internat. Zool. Cong
- 1 2 Stevens, N. M. (1908). "A study of the germ cells of certain diptera, with reference to the heterochromosomes and the phenomena of synapsis". Journal of Experimental Zoology. 5 (3): 359–374. doi:10.1002/jez.1400050304. ISSN 0022-104X.
- ↑ Fung, Jennifer C.; Marshall, Wallace F.; Dernburg, Abby; Agard, David A.; Sedat, John W. (1998). "Homologous Chromosome Pairing inDrosophila melanogasterProceeds through Multiple Independent Initiations". The Journal of Cell Biology. 141 (1): 5–20. doi:10.1083/jcb.141.1.5. ISSN 0021-9525.
- ↑ Gonzalez, Inma; Mateos-Langerak, Julio; Thomas, Aubin; Cheutin, Thierry; Cavalli, Giacomo (2014). "Identification of Regulators of the Three-Dimensional Polycomb Organization by a Microscopy-Based Genome-wide RNAi Screen". Molecular Cell. 54 (3): 485–499. doi:10.1016/j.molcel.2014.03.004. ISSN 1097-2765.
- ↑ Joyce, Eric F.; Williams, Benjamin R.; Xie, Tiao; Wu, C.-ting (2012). "Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen". PLoS Genetics. 8 (5): e1002667. doi:10.1371/journal.pgen.1002667. ISSN 1553-7404.
- ↑ Williams, B. R.; Bateman, J. R.; Novikov, N. D.; Wu, C.-T. (2007). "Disruption of Topoisomerase II Perturbs Pairing in Drosophila Cell Culture". Genetics. 177 (1): 31–46. doi:10.1534/genetics.107.076356. ISSN 0016-6731.
- ↑ Hartl, T. A.; Smith, H. F.; Bosco, G. (2008). "Chromosome Alignment and Transvection Are Antagonized by Condensin II". Science. 322 (5906): 1384–1387. doi:10.1126/science.1164216. ISSN 0036-8075.