Jaccard index

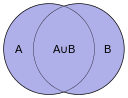
The Jaccard index, also known as the Jaccard similarity coefficient (originally coined coefficient de communauté by Paul Jaccard), is a statistic used for comparing the similarity and diversity of sample sets. The Jaccard coefficient measures similarity between finite sample sets, and is defined as the size of the intersection divided by the size of the union of the sample sets:
(If A and B are both empty, we define J(A,B) = 1.)
The Jaccard distance, which measures dissimilarity between sample sets, is complementary to the Jaccard coefficient and is obtained by subtracting the Jaccard coefficient from 1, or, equivalently, by dividing the difference of the sizes of the union and the intersection of two sets by the size of the union:
An alternate interpretation of the Jaccard distance is as the ratio of the size of the symmetric difference to the union.
This distance is a metric on the collection of all finite sets.[1][2]
There is also a version of the Jaccard distance for measures, including probability measures. If is a measure on a measurable space , then we define the Jaccard coefficient by , and the Jaccard distance by . Care must be taken if or , since these formulas are not well defined in that case.
The MinHash min-wise independent permutations locality sensitive hashing scheme may be used to efficiently compute an accurate estimate of the Jaccard similarity coefficient of pairs of sets, where each set is represented by a constant-sized signature derived from the minimum values of a hash function.
Similarity of asymmetric binary attributes
Given two objects, A and B, each with n binary attributes, the Jaccard coefficient is a useful measure of the overlap that A and B share with their attributes. Each attribute of A and B can either be 0 or 1. The total number of each combination of attributes for both A and B are specified as follows:
- represents the total number of attributes where A and B both have a value of 1.
- represents the total number of attributes where the attribute of A is 0 and the attribute of B is 1.
- represents the total number of attributes where the attribute of A is 1 and the attribute of B is 0.
- represents the total number of attributes where A and B both have a value of 0.
| A | |||
|---|---|---|---|
| 0 | 1 | ||
| B | 0 | ||
| 1 | |||
Each attribute must fall into one of these four categories, meaning that
The Jaccard similarity coefficient, J, is given as
The Jaccard distance, dJ, is given as
Difference with the Simple matching coefficient (SMC)
When used for binary attributes, the Jaccard index is very similar to the Simple matching coefficient. The main difference is that the SMC has the term in its numerator and denominator, whereas the Jaccard index does not. Thus, the SMC compares the number of matches with the entire set of the possible attributes, whereas the Jaccard index only compares it to the attributes that have been chosen by at least A or B.
In market basket analysis for example, the basket of two consumers who we wish to compare might only contain a small fraction of all the available products in the store, so the SMC would always return very small values compared to the Jaccard index. Using the SMC would then induce a bias by systematically considering, as more similar, two customers with large identical baskets compared to two customers with identical but smaller baskets; thus making the Jaccard index a better measure of similarity in that context.
In other contexts, where 0 and 1 carry equivalent information (symmetry), the SMC is a better measure of similarity. For example, vectors of demographic variables stored in dummies variables, such as gender, would be better compared with the SMC than with the Jaccard index since the impact of gender on similarity should be equal, independent of whether male is defined as a 0 and female as a 1 or the other way around. However, when we have symmetric dummy variables, one could replicate the behaviour of the SMC by splitting the dummies into two binary attributes (in this case, male and female), thus transforming them into asymmetric attributes, allowing the use of the Jaccard index without introducing the bias. By using this trick, the Jaccard index can be considered as making the SMC a fully redundant metric. The SMC remains however more computationally efficient in the case of symmetric dummy variables since it doesn't require adding extra dimensions.
In general, the Jaccard index can be considered as an indicator of local "similarity" while SMC evaluates "similarity" relative to the whole "universe". Similarity and dissimilarity must be understood in a relative sense. For example, if there are only 2 attributes (x,y), then A=(1,0) is intuitively very different from B=(0,1). However if there are 10 attributes in the "universe", A=(1,0,0,0,0,0,0,0,0,0) and B=(0,1,0,0,0,0,0,0,0) are not intuitively so different anymore. If the focus comes back to be just on A and B, the remaining 8 attributes are often considered as redundant. As a result, A and B are very different in a "local" sense (which the Jaccard Index measures efficiently), but less different in a "global" sense (which the SMC measures efficiently). From this point of view, the choice of using SMC or the Jaccard index comes down to more than just symmetry and asymmetry of information in the attributes. The distribution of sets in the defined "universe" and the nature of the problem to be modeled should also be considered.
The Jaccard index is also more general than the SMC and can be used to compare other data types than just vectors of binary attributes, such as Probability measures.
Generalized Jaccard similarity and distance
If and are two vectors with all real , then their Jaccard similarity coefficient is defined as
and Jaccard distance
With even more generality, if and are two non-negative measurable functions on a measurable space with measure , then we can define
where and are pointwise operators. Then Jaccard distance is
Then, for example, for two measurable sets , we have where and are the characteristic functions of the corresponding set.
Tanimoto similarity and distance
Various forms of functions described as Tanimoto similarity and Tanimoto distance occur in the literature and on the Internet. Most of these are synonyms for Jaccard similarity and Jaccard distance, but some are mathematically different. Many sources[3] cite an IBM Technical Report[4] as the seminal reference. The report is available from several libraries.
In "A Computer Program for Classifying Plants", published in October 1960,[5] a method of classification based on a similarity ratio, and a derived distance function, is given. It seems that this is the most authoritative source for the meaning of the terms "Tanimoto similarity" and "Tanimoto Distance". The similarity ratio is equivalent to Jaccard similarity, but the distance function is not the same as Jaccard distance.
Tanimoto's definitions of similarity and distance
In that paper, a "similarity ratio" is given over bitmaps, where each bit of a fixed-size array represents the presence or absence of a characteristic in the plant being modelled. The definition of the ratio is the number of common bits, divided by the number of bits set (i.e. nonzero) in either sample.
Presented in mathematical terms, if samples X and Y are bitmaps, is the ith bit of X, and are bitwise and, or operators respectively, then the similarity ratio is
If each sample is modelled instead as a set of attributes, this value is equal to the Jaccard coefficient of the two sets. Jaccard is not cited in the paper, and it seems likely that the authors were not aware of it.
Tanimoto goes on to define a "distance coefficient" based on this ratio, defined for bitmaps with non-zero similarity:
This coefficient is, deliberately, not a distance metric. It is chosen to allow the possibility of two specimens, which are quite different from each other, to both be similar to a third. It is easy to construct an example which disproves the property of triangle inequality.
Other definitions of Tanimoto distance
Tanimoto distance is often referred to, erroneously, as a synonym for Jaccard distance . This function is a proper distance metric. "Tanimoto Distance" is often stated as being a proper distance metric, probably because of its confusion with Jaccard distance.
If Jaccard or Tanimoto similarity is expressed over a bit vector, then it can be written as
where the same calculation is expressed in terms of vector scalar product and magnitude. This representation relies on the fact that, for a bit vector (where the value of each dimension is either 0 or 1) then and .
This is a potentially confusing representation, because the function as expressed over vectors is more general, unless its domain is explicitly restricted. Properties of do not necessarily extend to . In particular, the difference function does not preserve triangle inequality, and is not therefore a proper distance metric, whereas is.
There is a real danger that the combination of "Tanimoto Distance" being defined using this formula, along with the statement "Tanimoto Distance is a proper distance metric" will lead to the false conclusion that the function is in fact a distance metric over vectors or multisets in general, whereas its use in similarity search or clustering algorithms may fail to produce correct results.
Lipkus[1] uses a definition of Tanimoto similarity which is equivalent to , and refers to Tanimoto distance as the function . It is however made clear within the paper that the context is restricted by the use of a (positive) weighting vector such that, for any vector A being considered, . Under these circumstances, the function is a proper distance metric, and so a set of vectors governed by such a weighting vector forms a metric space under this function.
See also
- Sørensen similarity index
- Simple matching coefficient
- Most frequent k characters
- Hamming distance
- Dice's coefficient, which is equivalent: and
- Tversky index
- Correlation
- Mutual information, a normalized metricated variant of which is an entropic Jaccard distance.
Notes
- 1 2 Lipkus, Alan H (1999), "A proof of the triangle inequality for the Tanimoto distance", J Math Chem, 26 (1-3): 263–265
- ↑ Levandowsky, Michael; Winter, David (1971), "Distance between sets", Nature, 234 (5): 34–35, doi:10.1038/234034a0
- ↑ For example Qian, Huihuan; Wu, Xinyu; Xu, Yangsheng (2011). Intelligent Surveillance Systems. Springer. p. 161. ISBN 978-94-007-1137-2.
- ↑ Tanimoto, T. (17 Nov 1958). "An Elementary Mathematical theory of Classification and Prediction". Internal IBM Technical Report. 1957 (8?).
- ↑ Rogers, David J.; Tanimoto, Taffee T. (1960). "A Computer Program for Classifying Plants". Science. 132 (3434): 1115–1118. doi:10.1126/science.132.3434.1115.
References
- Tan, Pang-Ning; Steinbach, Michael; Kumar, Vipin (2005), Introduction to Data Mining, ISBN 0-321-32136-7.
- Jaccard, Paul (1901), "Étude comparative de la distribution florale dans une portion des Alpes et des Jura", Bulletin de la Société Vaudoise des Sciences Naturelles, 37: 547–579.
- Jaccard, Paul (1912), "The distribution of the flora in the alpine zone", New Phytologist, 11: 37–50, doi:10.1111/j.1469-8137.1912.tb05611.x.
External links
- Introduction to Data Mining lecture notes from Tan, Steinbach, Kumar
- SimMetrics a sourceforge implementation of Jaccard index and many other similarity metrics
- Web based tool for comparing texts using Jaccard coefficient
- A web based calculator for finding the Jaccard Coefficient
- Tutorial on how to calculate different similarities