Nucleotide pyrophosphatase/phosphodiesterase
| Nucleotide pyrophosphatase/phosphodiesterase (NPP) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
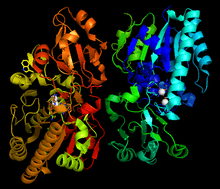 The overall dimeric structure of NPP in Xanthomonas axonopodis pv. citri str. 306 (Xac). This enzyme relies on the catalytic ability of 2 Zn2+ atoms in the catalytic core, which are shown in white.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 3.6.1.9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Nucleotide pyrophosphatase/phosphodiesterase (NPP) is a class of dimeric enzymes that catalyze the hydrolysis of phosphate diester bonds. NPP belongs to the alkaline phosphatase (AP) superfamily of enzymes.[2] Humans express seven known NPP isoforms,[3] some of which prefer nucleotide substrates, some of which prefer phospholipid substrates, and others of which prefer substrates that have not yet been determined.[4] In eukaryotes, most NPPs are located in the cell membrane and hydrolyze extracellular phosphate diesters to affect a wide variety of biological processes.[5][6] Bacterial NPP is thought to localize to the periplasm.[1]
Structure
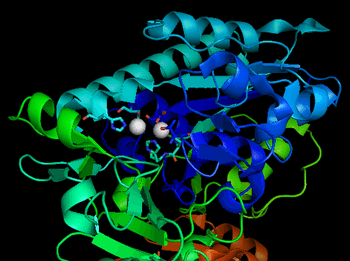
The catalytic site of NPP consists of a two-metal-ion (bimetallo) Zn2+ catalytic core. These Zn2+ catalytic components are thought to stabilize the transition state of the NPP phosphoryl transfer reaction.[7]
Mechanism
Overview
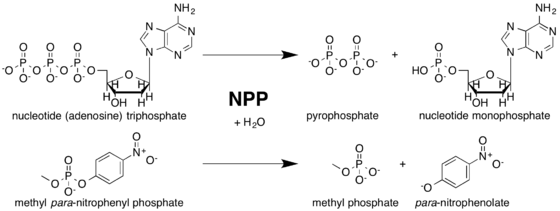
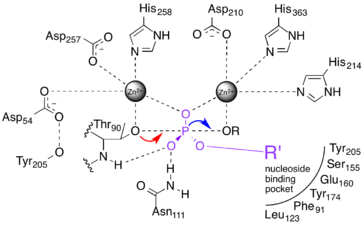
NPP catalyses the nucleophilic substitution of one ester bond on a phosphodiester substrate. It has a nucleoside binding pocket that excludes phospholipid substrates from the active site.[8] A threonine nucleophile has been identified through site-directed mutagenesis,[9][10][11] and the reaction inverts the stereochemistry of the phosphorus center.[12] The sequence of bond breakage and formation has yet to be resolved.
Ongoing Investigation
Three extreme possibilities have been proposed for the mechanism of NPP-catalyzed phosphoryl transfer. They are distinguished by the sequence in which bonds to phosphorus are made and broken. Though this phenomenon is subtle, it is important for understanding the physiological roles of AP superfamily enzymes, and also to molecular dynamic modeling.
Extreme mechanistic scenarios:
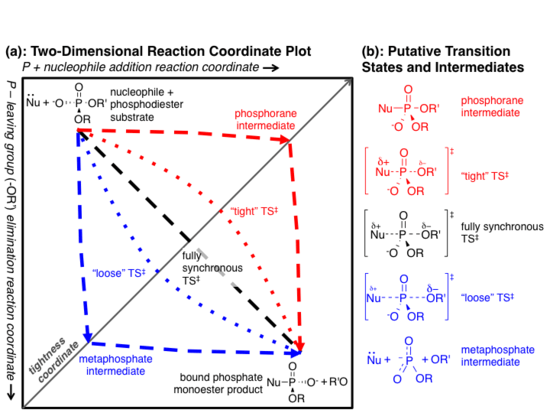
1) A two-step "dissociative" (elimination-addition or DN + AN) mechanism that proceeds via a trigonal metaphosphate intermediate.[13] This mechanism is represented by the red dashed lines in the figure at right.
2) A two-step "associative" (addition-elimination or AN + DN) mechanism that proceeds via a pentavalent phosphorane intermediate.[13] This is represented by the blue dashed lines in the figure at right.
3) A one-step fully synchronous mechanism analogous to SN2 substitution. Bond formation and breakage occur simultaneously and at the same rate. This is represented by the black dashed line in the figure at right.
The above three cases represent archetypes for the reaction mechanism, and the actual mechanism probably falls somewhere in between them.[13][14] The red and blue dotted lines in Fig. 2a represent more realistic "concerted" mechanisms in which addition and elimination overlap, but are not fully synchronous. The difference in initial rates of the two steps implies different charge distribution in the transition state (TS).
When the addition step occurs more quickly than elimination (an ANDN mechanism),[13] more positive charge develops on the nucleophile, and the transition state is said to be "tight."[1][14] Conversely, if elimination occurs more quickly than addition (DNAN), the transition state is considered "loose."
López-Canut et al. modeled substitution of a phosphodiester substrate using a hybrid quantum mechanics/molecular mechanics model.[14] Notably, the model predicted an ANDN concerted mechanism in aqueous solution, but a DNAN mechanism in the active site of Xac NPP.
Promiscuity
Although NPP primarily catalyzes phosphodiester hydrolysis, the enzyme will also catalyze the hydrolysis of phosphate monoesters, though to a much smaller extent. NPP preferentially hydrolyzes phosphate diesters over monoesters by factors of 102-106, depending on the identity of the diester substrate. This ability to catalyze a reaction with a secondary substrate is known as enzyme promiscuity,[1] and may have played a role in NPP’s evolutionary history.[15]
NPP’s promiscuity enables the enzyme to share substrates with alkaline phosphatase (AP), another member of the alkaline phosphate superfamily. Alkaline phosphatase primarily hydrolyzes phosphate monoester bonds, but interestingly it shows some promiscuity towards hydrolyzing phosphate diester bonds, making it a sort of opposite to NPP. The active sites of these two enzymes show marked similarities, namely in the presence of nearly superimposable Zn2+ bimetallo catalytic centers. In addition to the bimetallo core, AP also has an Mg2+ ion in its active site.[1]
Biological Function
NPPs have been implicated in several biological processes, including bone mineralization, purine nucleotide and insulin signaling, and cell differentiation and motility. They are generally regulated at the transcriptional level.[12]
Mammalian Isoforms
NPP1
NPP1 helps scavenge extracellular nucleotides in order to meet the high purine and pyrimidine requirements of dividing cells.[12] In T-cells, it may scavenge NAD+ from nearby dead cells as a source of adenosine.[16]
The pyrophosphate produced by NPP1 in bone cells is thought to serve as both a phosphate source for calcium phosphate deposition and as an inhibitory modulator of calcification.[17] NPP1 appears to be important for maintaining pyrophosphate/phosphate balance. Overactivity of the enzyme is associated with chondrocalcinosis, while deficiency correlates to pathological calcification.[6]
NPP1 inhibits the insulin receptor in vitro. In 2005, overexpression of the isoform was implicated in insulin resistance in mice.[18] It has been linked to insulin resistance and Type 2 diabetes in humans.[12]
NPP2
NPP2, known in humans as autotaxin, acts primarily in cell motility pathways. With its active site functioning, NPP2 promotes cellular migration at picomolar concentrations.[12] Soluble splice variants of NPP2 are thought to be important to cancer metastasis, and also show angiogenic properties in tumors.[6]
NPP3
NPP3 is probably a major contributor to nucleotide metabolism in the intestine and liver.[12]
Intestinal NPP3 would be involved in hydrolyzing food-derived nucleotides.[19]
The liver releases ATP and ADP into the bile to regulate bile secretion.[20] It subsequently reclaims adenosine via a pathway that probably contains NPP3.[21]
Evolution
NPP belongs to the alkaline phosphatase superfamily, which is a group of evolutionarily related enzymes that catalyze phosphoryl and sulfuryl transfer reactions. This group includes phosphomonoesterases, phosphodiesterases, phosphoglycerate mutases, phosphophenomutases, and sulfatases.[22]
References
- 1 2 3 4 5 6 7 Zalatan, JG; Fenn, TD; Brunger, AT; Herschlag, D (2006). "Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: Implications for mechanism and evolution". Biochemistry. 45 (32): 9788–803. doi:10.1021/bi060847t. PMID 16893180.
- ↑ Enzyme Promiscuity, Evolution, and Phosphoryl Transfer. Herschlag Lab. Stanford University, accessdate 1 March 2012.
- ↑ Terkeltaub, Robert (2006-06-01). "Physiologic and pathologic functions of the NPP nucleotide pyrophosphatase/phosphodiesterase family focusing on NPP1 in calcification". Purinergic Signalling. 2 (2): 371–377. doi:10.1007/s11302-005-5304-3. ISSN 1573-9538. PMC 2254483
 . PMID 18404477.
. PMID 18404477. - ↑ Pham, Truc Chi T.; Wanjala, Irene; Howard, Angela; Parrill, Abby L.; Baker, Daniel L. "Insights into the structure and function of lipid preferring Nucleotide PyrophosphotasePhosphodiesterase isoforms" 2011
- ↑ Stefan, Cristiana; Jansen, Silvia; Bollen, Mathieu (2005). "NPP-type ectophosphodiesterases: Unity in diversity". Trends in Biochemical Sciences. 30 (10): 542–50. doi:10.1016/j.tibs.2005.08.005. PMID 16125936.
- 1 2 3 Goding, James W.; Grobben, Bert; Slegers, Herman (2003). "Physiological and pathophysiological functions of the ecto-nucleotide pyrophosphatase/phosphodiesterase family". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1638: 1–19. doi:10.1016/S0925-4439(03)00058-9.
- ↑ Bobyr, Elena; Lassila, Jonathan K.; Wiersma-Koch, Helen I.; Fenn, Timothy D.; Lee, Jason J.; Nikolic-Hughes, Ivana; Hodgson, Keith O.; Rees, Douglas C.; et al. (2012). "High-Resolution Analysis of Zn2+ Coordination in the alkaline phosphatase Superfamily by EXAFS and X-ray Crystallography". Journal of Molecular Biology. 415 (1): 102–17. doi:10.1016/j.jmb.2011.10.040. PMC 3249517
 . PMID 22056344.
. PMID 22056344. - ↑ Stefan, C.; Gijsbers, R.; Stalmans, W.; Bollen, M. (1999-05-06). "Differential regulation of the expression of nucleotide pyrophosphatases/phosphodiesterases in rat liver". Biochimica et Biophysica Acta. 1450 (1): 45–52. doi:10.1016/s0167-4889(99)00031-2. ISSN 0006-3002. PMID 10231554.
- ↑ Belli, Sabina I.; Mercuri, Francesca A.; Sali, Adnan; Goding, James W. (1995-03-01). "Autophosphorylation of PC-1 (Alkaline Phosphodiesterase I/Nucleotide Pyrophosphatase) and Analysis of the Active Site". European Journal of Biochemistry. 228 (3): 669–676. doi:10.1111/j.1432-1033.1995.0669m.x. ISSN 1432-1033.
- ↑ Lee, Hoi Young; Clair, Timothy; Mulvaney, Peter T.; Woodhouse, Elisa C.; Aznavoorian, Sadie; Liotta, Lance A.; Stracke, Mary L. (1996-10-04). "Stimulation of Tumor Cell Motility Linked to Phosphodiesterase Catalytic Site of Autotaxin". Journal of Biological Chemistry. 271 (40): 24408–24412. doi:10.1074/jbc.271.40.24408. ISSN 0021-9258.
- ↑ Gijsbers, R.; Ceulemans, H.; Stalmans, W.; Bollen, M. (2001-01-12). "Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases". The Journal of Biological Chemistry. 276 (2): 1361–1368. doi:10.1074/jbc.M007552200. ISSN 0021-9258. PMID 11027689.
- 1 2 3 4 5 6 Bollen, M.; Gijsbers, R.; Ceulemans, H.; Stalmans, W.; Stefan, C. (2000-01-01). "Nucleotide pyrophosphatases/phosphodiesterases on the move". Critical Reviews in Biochemistry and Molecular Biology. 35 (6): 393–432. doi:10.1080/10409230091169249. ISSN 1040-9238. PMID 11202013.
- 1 2 3 4 Lassila, JK; Zalatan, JG; Herschlag, D (2011). "Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis". Annual Review of Biochemistry. 80: 669–702. doi:10.1146/annurev-biochem-060409-092741. PMC 3418923
 . PMID 21513457.
. PMID 21513457. - 1 2 3 López-Canut, Violeta; Roca, Maite; Bertrán, Juan; Moliner, Vicent; Tuñón, Iñaki (2010-05-26). "Theoretical study of phosphodiester hydrolysis in nucleotide pyrophosphatase/phosphodiesterase. Environmental effects on the reaction mechanism". Journal of the American Chemical Society. 132 (20): 6955–6963. doi:10.1021/ja908391v. ISSN 1520-5126. PMID 20429564.
- ↑ O'Brien, Patrick J.; Herschlag, Daniel (1999-04-01). "Catalytic promiscuity and the evolution of new enzymatic activities". Chemistry & Biology. 6 (4): R91–R105. doi:10.1016/S1074-5521(99)80033-7. ISSN 1074-5521. PMID 10099128.
- ↑ Deterre, P.; Gelman, L.; Gary-Gouy, H.; Arrieumerlou, C.; Berthelier, V.; Tixier, J.-M.; Ktorza, S.; Goding, J.; Schmitt, C.; Bismuth, G. (1996). "Coordinated regulation in human T cells of nucleotide-hydrolyzing ecto-enzymatic activities, including CD38 and PC-1. Possible role in the recycling of nicotinamide adenine dinucleotide metabolites.". The Journal of Immunology. PMID 8759717.
- ↑ Meyer, John L. (1984-05-15). "Can biological calcification occur in the presence of pyrophosphate?". Archives of Biochemistry and Biophysics. 231 (1): 1–8. doi:10.1016/0003-9861(84)90356-4.
- ↑ Dong, Hengjiang; Maddux, Betty A.; Altomonte, Jennifer; Meseck, Marcia; Accili, Domenico; Terkeltaub, Robert; Johnson, Kristen; Youngren, Jack F.; Goldfine, Ira D. (2005-02-01). "Increased Hepatic Levels of the Insulin Receptor Inhibitor, PC-1/NPP1, Induce Insulin Resistance and Glucose Intolerance". Diabetes. 54 (2): 367–372. doi:10.2337/diabetes.54.2.367. ISSN 0012-1797. PMID 15677494.
- ↑ Byrd, J. C.; Fearney, F. J.; Kim, Y. S. (1985). "Rat Intestinal Nucleotide-Sugar Pyrophosphatase". Journal of Biological Chemistry.
- ↑ Schlenker T, Romac JM, Sharara AI, Roman RM, Kim SJ, LaRusso N, Liddle RA, Fitz JG (1997). "Regulation of biliary secretion through apical purinergic receptors in cultured rat cholangiocytes". American Journal of Physiology - Gastrointestinal and Liver Physiology.
- ↑ Scott, L J; Delautier, D; Meerson, N R; Trugnan, G; Goding, J W; Maurice, M (1997-04-01). "Biochemical and molecular identification of distinct forms of alkaline phosphodiesterase I expressed on the apical and basolateral plasma membrane surfaces of rat hepatocytes". Hepatology. 25 (4): 995–1002. doi:10.1002/hep.510250434. ISSN 1527-3350.
- ↑ LóPez-Canut, Violeta; Roca, Maite; Bertrán, Juan; Moliner, Vicent; Tuñón, Iñaki (2010). "Theoretical Study of Phosphodiester Hydrolysis in Nucleotide Pyrophosphatase/Phosphodiesterase. Environmental Effects on the Reaction Mechanism". Journal of the American Chemical Society. 132 (20): 6955–63. doi:10.1021/ja908391v. PMID 20429564.