Penicillin binding proteins
| Penicillin-binding protein, transpeptidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PCN-bd_Tpept | ||||||||
| Pfam | PF00905 | ||||||||
| InterPro | IPR001460 | ||||||||
| |||||||||
| Penicillin-binding protein, dimerisation domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PBP_dimer | ||||||||
| Pfam | PF03717 | ||||||||
| InterPro | IPR005311 | ||||||||
| |||||||||

Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics (except for tabtoxinine-β-lactam, which inhibits glutamine synthetase) bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
Diversity
There are a large number of PBPs, usually several in each organism, and they are found as both membrane-bound and cytoplasmic proteins. For example, Spratt (1977) reports that six different PBPs are routinely detected in all strains of E. coli ranging in molecular weight from 40,000 to 91,000.[2] The different PBPs occur in different numbers per cell and have varied affinities for penicillin (see appendix). The PBPs are usually broadly classified into high-molecular-weight (HMW) and low-molecular-weight (LMW) categories.[3] Proteins that have evolved from PBPs occur in many higher organisms and include the mammalian LACTB protein.[4]
Function
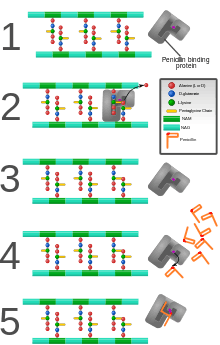
PBPs are all involved in the final stages of the synthesis of peptidoglycan, which is the major component of bacterial cell walls. Bacterial cell wall synthesis is essential to growth, cell division (thus reproduction) and maintaining the cellular structure in bacteria. Inhibition of PBPs leads to irregularities in cell wall structure such as elongation, lesions, loss of selective permeability, and eventual cell death and lysis.
PBPs have been shown to catalyze a number of reactions involved in the process of synthesizing cross-linked peptidoglycan from lipid intermediates and mediating the removal of D-alanine from the precursor of peptidoglycan. Purified enzymes have been shown to catalyze the following reactions: D-alanine carboxypeptidase, peptidoglycan transpeptidase, and peptidoglycan endopeptidase. In all bacteria that have been studied, enzymes have been shown to catalyze more than one of the above reactions.[2] The enzyme has a penicillin-insensitive transglycosylase N-terminal domain (involved in formation of linear glycan strands) and a penicillin-sensitive transpeptidase C-terminal domain (involved in cross-linking of the peptide subunits) and the serine at the active site is conserved in all members of the PBP family.[3]
Antibiotics
PBPs bind to β-lactam antibiotics because they are similar in chemical structure to the modular pieces that form the peptidoglycan.[5] When they bind to penicillin, the β-lactam amide bond is ruptured to form a covalent bond with the catalytic serine residue at the PBPs active site. This is an irreversible reaction and inactivates the enzyme.
There has been a great deal of research into PBPs because of their role in antibiotics and resistance. Bacterial cell wall synthesis and the role of PBPs in its synthesis is a very good target for drugs of selective toxicity because the metabolic pathways and enzymes are unique to bacteria.[6] Resistance to antibiotics has come about through overproduction of PBPs and formation of PBPs that have low affinity for penicillins (among other mechanisms such as lactamase production). Research on PBPs has led to the discovery of new semi-synthetic β-lactams, wherein altering the side-chains on the original penicillin molecule has increased the affinity of PBPs for penicillin, and, thus, increased effectiveness in bacteria with developing resistance.
Presence of the protein penicillin binding protein 2A (PBP2A) is responsible for the antibiotic resistance seen in methicillin-resistant Staphylococcus aureus (MRSA).
The β-lactam ring is a structure common to all β-lactam antibiotics.
See also
References
- ↑ Sainsbury, S., Bird, L., Rao, V., Shepherd, S.M., Stuart, D.I., Hunter, W.N., Owens, R.J., and Ren, J. (2011). "Crystal Structures of Penicillin-Binding Protein 3 from Pseudomonas aeruginosa: Comparison of Native and Antibiotic-Bound Forms". J.Mol.Biol. 405: 173–184. doi:10.1016/j.jmb.2010.10.024.
- 1 2 Spratt BG (1977). "Properties of the penicillin-binding proteins of Escherichia coli K12,.". Eur J Biochem. 72 (2): 341–52. doi:10.1111/j.1432-1033.1977.tb11258.x. PMID 319999.
- 1 2 Basu J, Chattopadhyay R, Kundu M, Chakrabarti P (1992). "Purification and partial characterization of a penicillin-binding protein from Mycobacterium smegmatis.". J Bacteriol. 174 (14): 4829–32. PMC 206282
 . PMID 1624470.
. PMID 1624470. - ↑ Peitsaro N, Polianskyte Z, Tuimala J, Pörn-Ares I, Liobikas J, Speer O, Lindholm D, Thompson J, Eriksson O (2008). "Evolution of a family of metazoan active-site-serine enzymes from penicillin-binding proteins: a novel facet of the bacterial legacy". MBC Evolutionary Biology. 8: 16. doi:10.1186/1471-2148-8-26. PMC 2266909
 . PMID 18226203.
. PMID 18226203. - ↑ Nguyen-Distèche M, Leyh-Bouille M, Ghuysen JM (1982). "Isolation of the membrane-bound 26 000-Mr penicillin-binding protein of Streptomyces strain K15 in the form of a penicillin-sensitive D-alanyl-D-alanine-cleaving transpeptidase.". Biochem J. 207 (1): 109–15. PMC 1153830
 . PMID 7181854.
. PMID 7181854. - ↑ Chambers HF (1999). "Penicillin-binding protein-mediated resistance in pneumococci and staphylococci.". J Infect Dis. 179 Suppl 2: S353–9. doi:10.1086/513854. PMID 10081507.