Reverse vaccinology
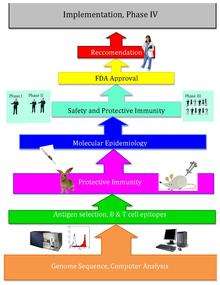
Reverse vaccinology is an improvement on vaccinology that employs bioinformatics, pioneered by Rino Rappuoli and first used against Serogroup B meningococcus.[1] Since then, it has been used on several other bacterial vaccines.[2]
Computational approach
The basic idea behind reverse vaccinology is that an entire pathogenic genome can be screened using bioinformatics approaches to find genes. Some of the traits that the genes are monitored for that may indicate antigenecity include genes that code for proteins with extracellular localization, signal peptides, and B-cell epitopes.[3] Next, those genes are filtered for desirable attributes that would make good vaccine targets such as outer membrane proteins. Once the candidates are identified, they are produced synthetically and are screened in animal models of the infection.
History
After Craig Venter published the genome of the first free-living organism in 1995, the genomes of other microorganisms became more readily available throughout the end of the twentieth century. Reverse vaccinology, designing vaccines using the pathogen’s sequenced genome, came from this new wealth of genomic information, as well as technological advances. Reverse vaccinology is much more efficient than traditional vaccinology, which requires growing large amounts of specific microorganisms as well as extensive wet lab tests.
In 2000, Rino Rappuoli and the J. Craig Venter Institute developed the first vaccine using Reverse Vaccinology against Serogroup B meningococcus. The J. Craig Venter institute and others then continued work on vaccines for A Streptococcus, B Streptococcus, Staphylococcus auereus, and Streptococcus pneumoniae. [4]
Reverse Vaccinology with Meningococcus B
Attempts at reverse vaccinology first began with Meningococcus B (MenB). Meningococcus B caused over 50% of meningococcal meningitis, and scientists had been unable to create a successful vaccine for the pathogen because of the bacterium’s unique structure. This bacterium’s polysaccharide shell is identical to that of a human self-antigen, but its surface proteins vary greatly; and the lack of information about the surface proteins caused developing a vaccine to be extremely difficult. As a result, Rino Rappuoli and other scientists turned towards bioinformatics to design a functional vaccine.[4]
Rappuoli and others at the J. Craig Venter Institute first sequenced the MenB genome. Then, they scanned the sequenced genome for potential antigens. They found over 600 possible antigens, which were tested by expression in Escherichia coli. The most universally applicable antigens were used in the prototype vaccines. Several proved to function successfully in mice, however, these proteins alone did not effectively interact with the human immune system due to not inducing a good immune response in order for the protection to be achieved. Later, by addition of outer membrane vesicles that contain lipopolysaccharides from the purification of blebs on gram negative cultures. The addition of this adjuvant (previously identified by using conventional vaccinology approaches) enhanced immune response to the level that was required. Later, the vaccine was proven to be safe and effective in adult humans.[4]
Subsequent Reverse Vaccinology Research
During the development of the MenB vaccine, scientists adopted the same Reverse Vaccinology methods for other bacterial pathogens. A Streptococcus and B Streptococcus vaccines were two of the first Reverse Vaccines created. Because those bacterial strains induce antibodies that react with human antigens, the vaccines for those bacteria needed to not contain homologies to proteins encoded in the human genome in order to not cause adverse reactions, thus establishing the need for genome-based Reverse Vaccinology.[4]
Later, Reverse Vaccinology was used to develop vaccines for antibiotic-resistant Staphylococcus aureus and Streptococcus pneumoniae [4]
Pros and cons
The major advantage for reverse vaccinology is finding vaccine targets quickly and efficiently. Traditional methods may take decades to unravel pathogens and antigens, diseases and immunity. However, In silico can be very fast, allowing to identify new vaccines for testing in only a few years.[5] The downside is that only proteins can be targeted using this process. Whereas, conventional vaccinology approaches can find other biomolecular targets such as polysaccharides.
Available Software
Though using bioinformatic technology to develop vaccines has become typical in the past ten years, general laboratories often do not have the advanced software that can do this. However, there are a growing number of programs making reverse vaccinology information more accessible. NERVE is one relatively new dataprocessing program. Though it must be downloaded and does not include all epitope predictions, it does help save some time by combining the computational steps of reverse vaccinology into one program. Vaxign, an even more comprehensive program, was created in 2008. Vaxign is web-based and completely public-access.[6]
Though Vaxign has been found to be extremely accurate and efficient, some scientists still utilize the online software RANKPEP for the peptide bonding predictions. Both Vaxign and RANKPEP employ PSSMs (Position Specific Scoring Matrices) when analyzing protein sequences or sequence alignments.[7]
Computer Aided bioinformatics projects are becoming extremely popular, as they help guide the laboratory experiments.[8]
Other Developments because of Reverse Vaccinology and Bioinformatics
- Reverse vaccinology has caused an increased focus on pathogenic biology.[4]
- Reverse vaccinology led to the discovery of pili in gram-positive pathogens such as A streptococcus, B streptococcus, and pneumococcus. Previously, all gram-positive bacteria were thought to not have any pili.[4]
- Reverse vaccinology also led to the discovery of factor G binding protein in meningococcus, which binds to complement factor H in humans. Binding to the complement factor H allows for meningococcus to grow in human blood while blocking alternative pathways. This model does not fit many animal species, which do not have the same complement factor H as humans, indicating differentiation of meningococcus between differing species.[4]
References
- ↑ Pizza et al. Identification of Vaccine Candidates Against Serogroup B Meningococcus by Whole-Genome Sequencing Science 2000 287:1816-1820
- ↑ Rappuoli, Rino. Reverse Vaccinology Current Opinion in Microbiology 2000, 3:445–450
- ↑ CH Woelk, et al. "Improving reverse vaccinology with a machine learning approach." Vaccine 29, no. 45 (n.d.): 8156-8164. Science Citation Index, EBSCOhost (accessed September 30, 2012).
- 1 2 3 4 5 6 7 8 Alessandro S, Rino R. Review: Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity [serial online]. n.d.;33:530-541. Available from: ScienceDirect, Ipswich, MA. Accessed September 30, 2012.
- ↑ Rappuoli, R. & A. Aderem. 2011. A 2020 Vision for vaccines against HIV, tuberculosis and malaria. Nature 473: 463.
- ↑ He Y, Xiang Z, Mobley H. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. Journal Of Biomedicine & Biotechnology [serial online]. 2010;Available from: CINAHL Plus with Full Text, Ipswich, MA. Accessed September 30, 2012.
- ↑ Reche PA, Glutting JP and Reinherz EL. Prediction of MHC Class I Binding Peptides Using Profile Motifs. Human Immunology 63, 701-709 (2002).
- ↑ Sandro V, Jennifer L. G, Francesco F, et al. Review: Computer-aided biotechnology: from immuno-informatics to reverse vaccinology. Trends In Biotechnology [serial online]. n.d.;26:190-200. Available from: ScienceDirect, Ipswich, MA. Accessed September 30, 2012.
External links
Profile of Rino Rappuoli: http://www.pnas.org/content/103/29/10831.full