Rsa RNA
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG.[1][2] Others, RsaOH-OX, were found thanks to an RNomic approach.[3] Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).[1]
RsaE
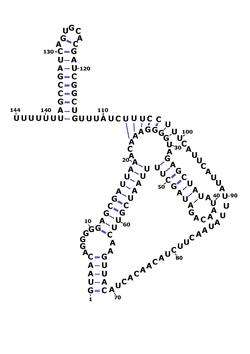
RsaE is found in other members of the Staphylococcus genus such as Staphylococcus epidermidis and Staphylococcus saprophyticus and is the only Rsa RNA to be found outside of this genus, in Macrococcus caseolyticus and Bacillus. In Bacillus subtilis, RsaE had previously been identified as ncr22.[6][7] RsaE is also consistently found downstream of PepF which codes for oligoendopeptidase F. The function of RsaE was discovered using gene knockout analysis and gene overexpression - it was found to regulate the expression of several enzymes involved in metabolism via antisense binding of their mRNA.[1][3]
RsaF
In S.aureus species RsaF is located in the same intergenic region as RsaE and overlaps with 3' end of RsaE by approximately 20bp. In contrast to RsaE, RsaF and its upstream gene have only been identified in S.aureus species.[1]
RsaK
RsaK is found in the leader sequence of glcA mRNA which encodes an enzyme involved in the glucose-specific phosphotransferase system. RsaK also contains a conserved ribonucleic antiterminator system, as recognised by GclT protein.[8]
RsaI
RsaOG[2] also renamed RsaI[1] is thought to fine-tune the regulation of toxin or invasion mechanisms in S. aureus via trans-acting mechanisms. Its secondary structure contains a pseudoknot formed between two highly conserved unpaired sequences.[4]
Expression patterns
RsaD, E H and I were found to be highly expressed in S. aureus. Expression levels of other Rsa RNAs varied under various environmental conditions, for example RsaC was induced by cold shock and RsaA is induced in response to osmotic stress.[1][2][3]
RsaE and RsaF genes overlap in S.aureus species but appear to have opposite expression patterns.[1] Transcriptional interference due to an overlap between a σA recognition motif and a potential σB binding site is proposed as a mechanism causing the differential expression of the two transcripts [1][9]
See also
References
- 1 2 3 4 5 6 7 8 Geissmann T, Chevalier C, Cros MJ, et al. (November 2009). "A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation". Nucleic Acids Res. 37 (21): 7239–57. doi:10.1093/nar/gkp668. PMC 2790875
 . PMID 19786493. Retrieved 2010-08-24.
. PMID 19786493. Retrieved 2010-08-24. - 1 2 3 Marchais A, Naville M, Bohn C, et al. (June 2009). "Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles". Genome Res. 19 (6): 1084–92. doi:10.1101/gr.089714.108. PMC 2694484
 . PMID 19237465.
. PMID 19237465. - 1 2 3 Bohn C, Rigoulay C, Chabelskaya S, et al. (2010). "Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism". Nucleic Acids Res. 38 (19): 6620–6636. doi:10.1093/nar/gkq462. PMC 2965222
 . PMID 20511587.
. PMID 20511587. - 1 2 Marchais A, Bohn C, Bouloc P, Gautheret D (March 2010). "RsaOG, a new staphylococcal family of highly transcribed non-coding RNA". RNA Biol. 7 (2): 116–9. doi:10.4161/rna.7.2.10925. PMID 20200491. Retrieved 2010-08-27.
- ↑ Darty K, Denise A, Ponty Y (August 2009). "VARNA: Interactive drawing and editing of the RNA secondary structure". Bioinformatics. 25 (15): 1974–5. doi:10.1093/bioinformatics/btp250. PMC 2712331
 . PMID 19398448. Retrieved 2010-07-12.
. PMID 19398448. Retrieved 2010-07-12. - ↑ Rasmussen S, Nielsen HB, Jarmer H (September 2009). "The transcriptionally active regions in the genome of Bacillus subtilis". Mol. Microbiol. 73 (6): 1043–57. doi:10.1111/j.1365-2958.2009.06830.x. PMC 2784878
 . PMID 19682248. Retrieved 2010-09-24.
. PMID 19682248. Retrieved 2010-09-24. - ↑ Irnov I, Sharma CM, Vogel J, Winkler WC (June 2010). "Identification of regulatory RNAs in Bacillus subtilis". Nucleic Acids Res. 38 (19): 6637–6651. doi:10.1093/nar/gkq454. PMC 2965217
 . PMID 20525796. Retrieved 2010-09-24.
. PMID 20525796. Retrieved 2010-09-24. - ↑ Langbein I, Bachem S, Stülke J (November 1999). "Specific interaction of the RNA-binding domain of the bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT". J. Mol. Biol. 293 (4): 795–805. doi:10.1006/jmbi.1999.3176. PMID 10543968. Retrieved 2010-08-27.
- ↑ Shearwin KE, Callen BP, Egan JB (June 2005). "Transcriptional interference--a crash course". Trends Genet. 21 (6): 339–45. doi:10.1016/j.tig.2005.04.009. PMC 2941638
 . PMID 15922833.
. PMID 15922833.
Further reading
- Pichon C, Felden B (October 2005). "Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains". Proc. Natl. Acad. Sci. U.S.A. 102 (40): 14249–54. doi:10.1073/pnas.0503838102. PMC 1242290
 . PMID 16183745. Retrieved 2010-08-24.
. PMID 16183745. Retrieved 2010-08-24. - Altuvia S (June 2007). "Identification of bacterial small non-coding RNAs: experimental approaches". Curr. Opin. Microbiol. 10 (3): 257–61. doi:10.1016/j.mib.2007.05.003. PMID 17553733. Retrieved 2010-08-24.
- Morfeldt E, Taylor D, von Gabain A, Arvidson S (September 1995). "Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII". EMBO J. 14 (18): 4569–77. PMC 394549
 . PMID 7556100.
. PMID 7556100. - Roberts C, Anderson KL, Murphy E, et al. (April 2006). "Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives". J. Bacteriol. 188 (7): 2593–603. doi:10.1128/JB.188.7.2593-2603.2006. PMC 1428411
 . PMID 16547047. Retrieved 2010-08-24.
. PMID 16547047. Retrieved 2010-08-24. - Anderson KL, Roberts C, Disz T, et al. (October 2006). "Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover". J. Bacteriol. 188 (19): 6739–56. doi:10.1128/JB.00609-06. PMC 1595530
 . PMID 16980476. Retrieved 2010-08-24.
. PMID 16980476. Retrieved 2010-08-24. - Huntzinger E, Boisset S, Saveanu C, et al. (February 2005). "Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression". EMBO J. 24 (4): 824–35. doi:10.1038/sj.emboj.7600572. PMC 549626
 . PMID 15678100.
. PMID 15678100. - Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S (October 1993). "Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule". EMBO J. 12 (10): 3967–75. PMC 413679
 . PMID 7691599.
. PMID 7691599. - Bohn C, Rigoulay C, Bouloc P (February 2007). "No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus". BMC Microbiol. 7: 10. doi:10.1186/1471-2180-7-10. PMC 1800855
 . PMID 17291347.
. PMID 17291347.