White Collar-1
| White Collar-1 | |
|---|---|
| Identifiers | |
| Organism | |
| Symbol | wc-1 |
| Entrez | 3875924 |
| RefSeq (mRNA) | XM_011396849 |
| RefSeq (Prot) | XP_011395151 |
| UniProt | Q01371 |
White Collar-1 (wc-1) is a gene in Neurospora crassa encoding the protein WC-1 (127 kDa).[1][2] WC-1 has two separate roles in the cell. First, it is the primary photoreceptor for Neurospora[3] and the founding member of the class of principle blue light photoreceptors in all of the fungi.[4] Second, it is necessary for regulating circadian rhythms in FRQ. It is a key component of a circadian molecular pathway that regulates many behavioral activities, including conidiation.[5][6] WC-1 and WC-2, an interacting partner of WC-1, comprise the White Collar Complex (WCC) that is involved in the Neurospora circadian clock. WCC is a complex of nuclear transcription factor proteins, and contains transcriptional activation domains, PAS domains, and zinc finger DNA-binding domains (GATA).[7] WC-1 and WC-2 heterodimerize through their PAS domains to form the White Collar Complex (WCC).[8][9] While mainly identified and studied in fungi, members of the White Collar-1 gene family have also been found to be expressed in bovine lymphocytes.[10]
Discovery
The Neurospora circadian clock was discovered in 1959, when Pittendrigh et al. first described timing patterns in the asexual development of spores.[11] They noticed that in the region of the growing front, mycelia laid down between the late night to early morning formed aerial hyphae, whereas those laid down at other times did not.[11][12] This aerial growth pattern at subjective circadian times served as tentative support for the presence of circadian oscillators.
White Collar-1 was described in the 1960s by geneticists who saw a strain of Neurospora which the mycelia were albino, but the conidia were normally pigmented.[13]
The mutant gene was designated white collar (wc), for the white coloration of the mycelia below the pigmented conidia on agar slants.[14] The gene wc-1 was classified during the mapping of the chromosomal loci of Neurospora crassa (1982 by Perkins et al.).[15]
Protein structure
WC-1 and WC-2 are transcription factors encoded by the genes wc-1 and wc-2. Zinc finger DNA-binding domains (GATA) allow WC-1 and WC-2 to bind to DNA and act as transcription factors.[1][8]
Both WC-1 and WC-2 have PAS domains that allow them to bind to other proteins.[11][16] WC-1 and WC-2 typically heterodimerize in vivo to form the White Collar Complex (WCC), which acts as a transcription factor complex. In vitro, WC-1 can also homodimerize with itself and heterodimerize with other PAS proteins.[11]
Protein sequencing has revealed WC-1 to also contain a LOV domain, a chromophore-binding peptide region.[2][17] This binding site is highly conserved, and is sequentially similar to the chromophore-binding domains of phototropins in plants.[18]
WC-1 and WC-2 bind to the promoter elements of the genes that they transcriptionally activate.[3][11]
Function in the circadian clock
Photoreception
WC-1 has been shown to be a blue-light photoreceptor, and is a necessary component of the Neurospora light-induced response pathway.[3] Genetic screens of light-insensitive Neurospora mutant strains have repeatedly demonstrated abnormalities in the wc-1 gene.[17] In functional Neurospora, the WC-1 LOV domain binds to the flavin adenine dinucleotide (FAD) chromophore,[3][19] which is responsible for the conversion of light to mechanical energy. FAD displays maximum absorption of light at 450 nm,[20] thus explaining WC-1's maximal sensitivity to blue light.
Light-induced responses are completely eliminated in WC-1 LOV knockout Neurospora mutants, although WC-1's transcription activation role persists in the dark.[17] WC-1 is widely conserved among fungi where it appears to be the principle blue light photoreceptor for the entire kingdom.[4]
Circadian regulation
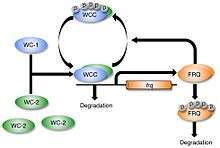
The White Collar Complex (WCC), the heterodimer of WC-1 and WC-2, acts as a positive element in the circadian clock. WCC serves as an activator of frq gene transcription by binding to two DNA promotor elements in the nucleus: the Clock box (C box) and the Proximal Light-Response Element (PLRE). PLRE is required for maximal light induction, while the C box is required for both maximal light induction and maintaining circadian rhythmicity in constant darkness.[11][23]
In addition, light-activated WCC is shown to induce the transcription of VIVID, a small flavin-binding blue-light photoreceptor that is required for adaptation to light-induced responses in the transcription of light-induced genes, including wc-1 and frq.[24][25][26] VIVID is a negative regulator of light responses, although its mechanism is not yet known.[27]
As part of the transcription-translation negative feedback-loop (TTFL), FRQ protein enters the nucleus and interacts with FRQ-interacting RNA Helicase (FRH) to promote the repression of WCC activity. This FRQ–FRH complex is suggested to recruit protein kinases such as casein kinase (CK I) and CK II to phosphorylate WCC.[28] The phosphorylation of the WCC stabilizes WCC, preventing it from binding and activating frq transcription. Protein phosphatases PP2A and PP4 are known to counterbalance kinase activity and support the reactivation and nuclear entry of WCC.[29]
FRQ has also been shown to interact with WC-2 in vitro, and a partial loss-of-function allele of wc-2 yields Neurospora with a long period length and altered temperature compensation, which is a key characteristic of circadian pacemakers.[11][30]
Only WC-1 is required for transient light-induction, but both WC-1 and WC-2 are required for the circadian clock to run.[11][31] Because the core of the clock is based on a rhythmic expression of frq, the acute light-induction pathway provides a direct way to reset the clock. Mammalian clocks can be reset through a similar mechanism, via the light-induction of the mammalian per genes within the SCN.[32]
Several WC-1 mutants are known. The rhy-2 mutation was localized to the polyglutamine region of the WC-1 gene product. The rhy-2 mutant is arrhythmic with regard to conidiation in constant darkness, but rhythmic in a light-dark cycle. Rhy-2 is also only weakly sensitive to light, suggesting that the polyglutamine region may be essential for both clock function and light sensing in Neurospora.[33]
See also
References
- 1 2 Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G (Apr 1996). "White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein". The EMBO Journal. 15 (7): 1650–7. PMID 8612589.
- 1 2 Lee K, Loros JJ, Dunlap JC (Jul 2000). "Interconnected feedback loops in the Neurospora circadian system". Science. 289 (5476): 107–110. doi:10.1126/science.289.5476.107. PMID 10884222.
- 1 2 3 4 Froehlich AC, Liu Y, Loros JJ, Dunlap JC (Aug 2002). "White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter". Science. 297 (5582): 815–9. doi:10.1126/science.1073681. PMID 12098706.
- 1 2 Dunlap JC, Loros JJ (Dec 2006). "How fungi keep time: circadian system in Neurospora and other fungi". Current Opinion in Microbiology. 9 (6): 579–87. doi:10.1016/j.mib.2006.10.008. PMID 17064954.
- ↑ Baker CL, Loros JJ, Dunlap JC (Jan 2012). "The circadian clock of Neurospora crassa". FEMS Microbiology Reviews. 36 (1): 95–110. doi:10.1111/j.1574-6976.2011.00288.x. PMID 21707668.
- 1 2 Crosthwaite SK, Dunlap JC, Loros JJ (May 1997). "Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity". Science. 276 (5313): 763–9. doi:10.1126/science.276.5313.763. PMID 9115195.
- ↑ Fungal Genomics. Elsevier. 2003.
- 1 2 Lee K, Dunlap JC, Loros JJ (Jan 2003). "Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa". Genetics. 163 (1): 103–14. PMC 1462414
 . PMID 12586700.
. PMID 12586700. - ↑ Talora C, Franchi L, Linden H, Ballario P, Macino G (Sep 1999). "Role of a white collar-1-white collar-2 complex in blue-light signal transduction". The EMBO Journal. 18 (18): 4961–8. doi:10.1093/emboj/18.18.4961. PMC 1171567
 . PMID 10487748.
. PMID 10487748. - ↑ Wijngaard PL, MacHugh ND, Metzelaar MJ, Romberg S, Bensaid A, Pepin L, Davis WC, Clevers HC (Apr 1994). "Members of the novel WC1 gene family are differentially expressed on subsets of bovine CD4-CD8- gamma delta T lymphocytes". Journal of Immunology. 152 (7): 3476–82. PMID 7511649.
- 1 2 3 4 5 6 7 8 Dunlap JC (Jan 1999). "Molecular bases for circadian clocks". Cell. 96 (2): 271–90. doi:10.1016/S0092-8674(00)80566-8. PMID 9988221.
- ↑ Pittendrigh CS, Bruce VG, Rosensweig NS, Rubin ML (Jul 1959). "Growth Patterns in Neurospora: A Biological Clock in Neurospora". Nature. 184 (4681): 169–170. doi:10.1038/184169a0.
- ↑ Perkins DD, Newmeyer D, Taylor CW, Bennett DC (1969). "New markers and map sequences in Neurospora crassa, with a description of mapping by duplication coverage, and of multiple translocation stocks for testing linkage". Genetica. 40 (3): 247–78. doi:10.1007/BF01787357. PMID 5367562.
- ↑ Harding RW, Shropshire W (1980). "Photocontrol of Carotenoid Biosynthesis". Annual Review of Plant Physiology. 31 (1): 217–238. doi:10.1146/annurev.pp.31.060180.001245.
- ↑ Perkins DD, Radford A, Newmeyer D, Björkman M (Dec 1982). "Chromosomal loci of Neurospora crassa". Microbiological Reviews. 46 (4): 426–570. PMID 6219280.
- ↑ Talora C, Franchi L, Linden H, Ballario P, Macino G (Sep 1999). "Role of a white collar-1-white collar-2 complex in blue-light signal transduction". The EMBO Journal. 18 (18): 4961–4968. doi:10.1093/emboj/18.18.4961. PMC 1171567
 . PMID 10487748.
. PMID 10487748. - 1 2 3 Liu Y, He Q, Cheng P (Oct 2003). "Photoreception in Neurospora: a tale of two White Collar proteins". Cellular and Molecular Life Sciences. 60 (10): 2131–2138. doi:10.1007/s00018-003-3109-5. PMID 14618260.
- ↑ He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y (Aug 2002). "White collar-1, a DNA binding transcription factor and a light sensor". Science. 297 (5582): 840–3. doi:10.1126/science.1072795. PMID 12098705.
- ↑ Kritskiĭ MS, Belozerskaia TA, Sokolovskiĭ VIu, Filippovich SIu (Jul–Aug 2005). "[Photoreceptor apparatus of a fungus Neurospora crassa]". Molekuliarnaia Biologiia (in Russian). 39 (4): 602–617. PMID 16083009.
- ↑ Lewis JA, Escalante-Semerena JC (Aug 2006). "The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate". Journal of Bacteriology. 188 (15): 5479–5486. doi:10.1128/JB.00514-06. PMID 16855237.
- ↑ Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC (May 2009). "Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock". Molecular Cell. 34 (3): 354–63. doi:10.1016/j.molcel.2009.04.023. PMID 19450533.
- ↑ Hurley J, Loros JJ, Dunlap JC (2015). "Dissecting the mechanisms of the clock in Neurospora". Methods in Enzymology. 551: 29–52. doi:10.1016/bs.mie.2014.10.009. PMID 25662450.
- ↑ Gooch VD, Johnson AE, Larrondo LF, Loros JJ, Dunlap JC (Feb 2014). "Bright to dim oscillatory response of the Neurospora circadian oscillator". Journal of Biological Rhythms. 29 (1): 49–59. doi:10.1177/0748730413517983. PMC 4083493
 . PMID 24492882.
. PMID 24492882. - ↑ Schwerdtfeger C, Linden H (Sep 2003). "VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation". The EMBO Journal. 22 (18): 4846–4855. doi:10.1093/emboj/cdg451. PMID 12970196.
- ↑ Heintzen C, Loros JJ, Dunlap JC (Feb 2001). "The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting". Cell. 104 (3): 453–64. doi:10.1016/s0092-8674(01)00232-x. PMID 11239402.
- ↑ Schwerdtfeger C, Linden H (Feb 2001). "Blue light adaptation and desensitization of light signal transduction in Neurospora crassa". Molecular Microbiology. 39 (4): 1080–7. doi:10.1046/j.1365-2958.2001.02306.x. PMID 11251826.
- ↑ Brunner M, Káldi K (Apr 2008). "Interlocked feedback loops of the circadian clock of Neurospora crassa". Molecular Microbiology. 68 (2): 255–262. doi:10.1111/j.1365-2958.2008.06148.x. PMID 18312266.
- ↑ Shi M, Collett M, Loros JJ, Dunlap JC (Feb 2010). "FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock". Genetics. 184 (2): 351–361. doi:10.1534/genetics.109.111393.
- ↑ Cha J, Zhou M, Liu Y (2015). "Methods to study molecular mechanisms of the Neurospora circadian clock". Methods in Enzymology. 551: 137–151. doi:10.1016/bs.mie.2014.10.002.
- ↑ Wang B, Kettenbach AN, Gerber SA, Loros JJ, Dunlap JC (Sep 2014). "Neurospora WC-1 recruits SWI/SNF to remodel frequency and initiate a circadian cycle". PLoS Genetics. 10 (9): e1004599. doi:10.1371/journal.pgen.1004599.
- ↑ Crosthwaite SK, Loros JJ, Dunlap JC (Jun 1995). "Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript". Cell. 81 (7): 1003–1012. doi:10.1016/s0092-8674(05)80005-4. PMID 7600569.
- ↑ Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H (Dec 1997). "Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript". Cell. 91 (7): 1043–1053. doi:10.1016/s0092-8674(00)80494-8. PMID 9428526.
- ↑ Toyota K, Onai K, Nakashima H (Sep 2002). "A new wc-1 mutant of Neurospora crassa shows unique light sensitivity in the circadian conidiation rhythm". Molecular Genetics and Genomics. 268 (1): 56–61. doi:10.1007/s00438-002-0722-1. PMID 12242499.