16S ribosomal RNA
16S ribosomal RNA (or 16S rRNA) is the component of the 30S small subunit of a prokaryotic ribosome that binds to the Shine-Dalgarno sequence. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene.[2] Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenies.[3]
Multiple sequences of 16S rRNA can exist within a single bacterium.[4]
Functions
It has several functions:
- Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins.
- The 3' end contains the anti-Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3'-end of 16S RNA binds to the proteins S1 and S21 known to be involved in initiation of protein synthesis; RNA-protein cross-linking by A.P. Czernilofsky et al. (FEBS Lett. Vol 58, pp 281–284, 1975).
- Interacts with 23S, aiding in the binding of the two ribosomal subunits (50S+30S)
- Stabilizes correct codon-anticodon pairing in the A site, via a hydrogen bond formation between the N1 atom of Adenine (see image of Purine chemical structure) residues 1492 and 1493 and the 2'OH group of the mRNA backbone
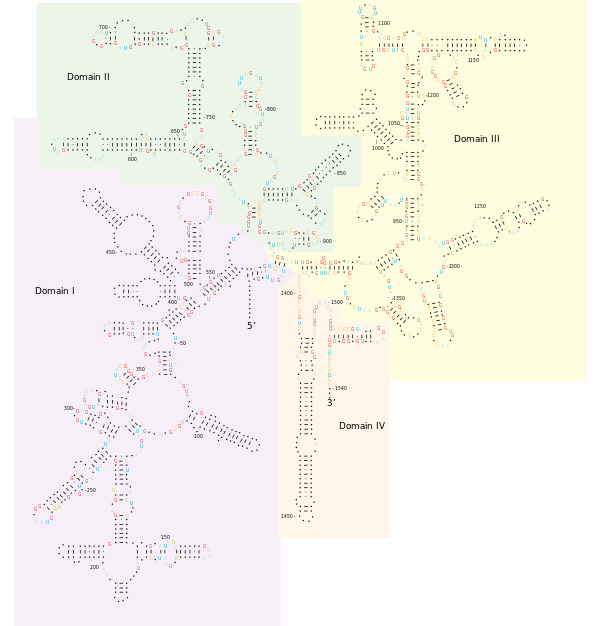
Structure
Universal primers
The 16S rRNA gene is used for phylogenetic studies[5] as it is highly conserved between different species of bacteria and archaea.[6] Carl Woese pioneered this use of 16S rRNA.[2] Some (hyper)thermophilic archaea (i.e. order Thermoproteales) contain 16S rRNA gene introns that are located in highly conserved regions and can impact the annealing of "universal" primers.[7] Mitochondrial and chloroplastic rRNA are also amplified.
The most common primer pair was devised by Weisburg et al.[5] and is currently referred to as 27F and 1492R; however, for some applications shorter amplicons may be necessary for example for 454 sequencing with Titanium chemistry (500-ish reads are ideal) the primer pair 27F-534R covering V1 to V3.[8] Often 8F is used rather than 27F. The two primers are almost identical, but 27F has an M instead of a C. AGAGTTTGATCMTGGCTCAG compared with 8F.[9]
| Primer name | Sequence (5'-3') | Reference |
|---|---|---|
| 8F | AGA GTT TGA TCC TGG CTC AG | [10][11] |
| U1492R | GGT TAC CTT GTT ACG ACT T | same as above |
| 928F | TAA AAC TYA AAK GAA TTG ACG GG | [12] |
| 336R | ACT GCT GCS YCC CGT AGG AGT CT | as above |
| 1100F | YAA CGA GCG CAA CCC | |
| 1100R | GGG TTG CGC TCG TTG | |
| 337F | GAC TCC TAC GGG AGG CWG CAG | |
| 907R | CCG TCA ATT CCT TTR AGT TT | |
| 785F | GGA TTA GAT ACC CTG GTA | |
| 805R | GAC TAC CAG GGT ATC TAA TC | |
| 533F | GTG CCA GCM GCC GCG GTA A | |
| 518R | GTA TTA CCG CGG CTG CTG G | |
| 27F | AGA GTT TGA TCM TGG CTC AG | [13] |
| 1492R | CGG TTA CCT TGT TAC GAC TT | as above |
PCR applications
In addition to highly conserved primer binding sites, 16S rRNA gene sequences contain hypervariable regions that can provide species-specific signature sequences useful for identification of bacteria.[14][15] As a result, 16S rRNA gene sequencing has become prevalent in medical microbiology as a rapid and cheap alternative to phenotypic methods of bacterial identification.[16] Although it was originally used to identify bacteria, 16S sequencing was subsequently found to be capable of reclassifying bacteria into completely new species,[17] or even genera.[18][19] It has also been used to describe new species that have never been successfully cultured.[20][21]
16S ribosomal databases
The 16S rRNA gene is used as the standard for classification and identification of microbes, because it is present in most microbes and shows proper changes. Type strains of 16S rRNA gene sequences for most bacteria and archaea are available on public databases such as NCBI. However, the quality of the sequences found on these databases are often not validated. Therefore, secondary databases that collect only 16S rRNA sequences are widely used. The most frequently used databases are listed below:
EzTaxon-e
http://eztaxon-e.ezbiocloud.net/ The EzTaxon-e database is an extension of the original EzTaxon database. It contains comprehensive 16S rRNA gene sequences of taxa with valid names as well as sequences of uncultured taxa. EzTaxon-e contains complete hierarchical taxonomic structure (from phylum rank to species rank) for the domain of bacteria and archaea.[22]
Ribosomal Database Project
http://rdp.cme.msu.edu/ The Ribosomal Database Project (RDP) is a curated database that offers ribosome data along with related programs and services. The offerings include phylogenetically ordered alignments of ribosomal RNA (rRNA) sequences, derived phylogenetic trees, rRNA secondary structure diagrams and various software packages for handling, analyzing and displaying alignments and trees. The data are available via ftp and electronic mail. Certain analytic services are also provided by the electronic mail server.[23]
SILVA
SILVA provides comprehensive, quality checked and regularly updated datasets of aligned small (16S/18S, SSU) and large subunit (23S/28S, LSU) ribosomal RNA (rRNA) sequences for all three domains of life as well as a suite of search, primer-design and alignment tools (Bacteria, Archaea and Eukarya).[24] (Warning: the latest version with taxonomies, SILVA_123.1_SSURef_Nr99_tax, contains errors in the taxonomy because the species name is taken from the source and the taxonomy, up to genus, is found by similarity, e.g. like a pseudomonas lineage up to genus with a cricket species: Pseudomonas;Teleogryllus commodus.)
Greengenes
Greengenes is a quality controlled, comprehensive 16S reference database and taxonomy based on a de novo phylogeny that provides standard operational taxonomic unit sets. The official home page for the site is http://greengenes.secondgenome.com, and is licensed under the Creative Commons BY-SA 3.0 license.[25][26]
References
- ↑ Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A (2000). "Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution". Cell. 102 (5): 615–23. doi:10.1016/S0092-8674(00)00084-2. PMID 11007480.
- 1 2 Woese, C. R.; G. E. Fox (1977-11-01). "Phylogenetic structure of the prokaryotic domain: The primary kingdoms". Proceedings of the National Academy of Sciences. 74 (11): 5088–5090. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. ISSN 0027-8424. PMC 432104
 . PMID 270744.
. PMID 270744.
- ↑ Woese, Carl R.; Kandler, O; Wheelis, M (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proc Natl Acad Sci USA. 87 (12): 4576–9. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159
 . PMID 2112744.
. PMID 2112744. - ↑ Case RJ, Boucher Y, Dahllöf I, Holmström C, Doolittle WF, Kjelleberg S (January 2007). "Use of 16S rRNA and rpoB Genes as Molecular Markers for Microbial Ecology Studies". Appl. Environ. Microbiol. 73 (1): 278–88. doi:10.1128/AEM.01177-06. PMC 1797146
 . PMID 17071787.
. PMID 17071787. - 1 2 Weisburg WG, Barns SM, Pelletier DA, Lane DJ (January 1991). "16S ribosomal DNA amplification for phylogenetic study". J Bacteriol. 173 (2): 697–703. PMC 207061
 . PMID 1987160.
. PMID 1987160. - ↑ Coenye T, Vandamme P (November 2003). "Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes". FEMS Microbiol. Lett. 228 (1): 45–49. doi:10.1016/S0378-1097(03)00717-1. PMID 14612235.
- ↑ Jay ZJ, Inskeep WP (July 2015). "The distribution, diversity, and importance of 16S rRNA gene introns in the order Thermoproteales.". BiologyDirect. 10 (35). doi:10.1186/s13062-015-0065-6. PMID 26156036.
- ↑ http://www.hmpdacc.org/tools_protocols.php#sequencing
- ↑ Primers, 16S ribosomal DNA - François Lutzoni's Lab
- ↑ Eden PA, Schmidt TM, Blakemore RP, Pace NR (1991). "Phylogenetic Analysis of Aquaspirillum magnetotacticum Using Polymerase Chain Reaction-Amplified 16S rRNA-Specific DNA". Int J Syst Bacteriol. 41 (2): 324–325. doi:10.1099/00207713-41-2-324. PMID 1854644.
- ↑ Universal Bacterial Identification by PCR and DNA Sequencing of 16S rRNA Gene. PCR for Clinical Microbiology, 2010, Part 3, 209-214
- ↑ Weidner S, Arnold W, Pühler A (1996). "Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes" (PDF). Appl Environ Microbiol. 62 (3): 766–71.
- ↑ Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L. R.; Fields, M. W. (2006). "Microbial Diversity in Water and Sediment of Lake Chaka, an Athalassohaline Lake in Northwestern China". Applied and Environmental Microbiology. 72 (6): 3832–3845. doi:10.1128/AEM.02869-05. PMC 1489620
 . PMID 16751487.
. PMID 16751487. - ↑ Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmao, L.; Amorim, A. (4 October 2010). "Identification of species by multiplex analysis of variable-length sequences". Nucleic Acids Research. 38 (22): e203–e203. doi:10.1093/nar/gkq865.
- ↑ Kolbert, CP; Persing, DH (June 1999). "Ribosomal DNA sequencing as a tool for identification of bacterial pathogens". Current Opinion in Microbiology. 2 (3): 299–305. doi:10.1016/S1369-5274(99)80052-6. PMID 10383862.
- ↑ J. E. Clarridge III (2004). "Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases". Clin Microbiol Rev. 17 (4): 840–862. doi:10.1128/CMR.17.4.840-862.2004. PMC 523561
 . PMID 15489351.
. PMID 15489351. - ↑ Lu T, Stroot PG, Oerther DB (2009). "Reverse Transcription of 16S rRNA To Monitor Ribosome-Synthesizing Bacterial Populations in the Environment". Appl Environ Microbiol. 75 (13): 4589–4598. doi:10.1128/AEM.02970-08. PMC 2704851
 . PMID 19395563.
. PMID 19395563. - ↑ Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991). "16S ribosomal DNA amplification for phylogenetic study". J Bacteriol. 173 (2): 697–703. PMC 207061
 . PMID 1987160.
. PMID 1987160. - ↑ Brett PJ, DeShazer D, Woods DE (1998). "Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species". Int J Syst Bacteriol. 48 (1): 317–320. doi:10.1099/00207713-48-1-317. PMID 9542103.
- ↑ Schmidt TM, Relman DA (1994). "Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences". Methods Enzymol. Methods in Enzymology. 235: 205–22. doi:10.1016/0076-6879(94)35142-2. ISBN 978-0-12-182136-4. PMID 7520119.
- ↑ Gray JP, Herwig RP (1996). "Phylogenetic analysis of the bacterial communities in marine sediments". Appl Environ Microbiol. 62 (11): 4049–59. PMC 168226
 . PMID 8899989.
. PMID 8899989. - ↑ Chun, J.; Lee, J.-H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B. K.; Lim, Y. W. (2007). "EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences". Int J Syst Evol Microbiol. 57: 2259–2261. doi:10.1099/ijs.0.64915-0.
- ↑ Larsen N, Olsen GJ, Maidak BL, McCaughey MJ, Overbeek R, Macke TJ, Marsh TL, Woese CR. (1993) The ribosomal database project. Nucleic Acids Res. Jul 1;21(13):3021-3.
- ↑ Elmar Pruesse, Christian Quast, Katrin Knittel, Bernhard M. Fuchs, Wolfgang Ludwig, Jörg Peplies, Frank Oliver Glöckner (2007) Nucleic Acids Res. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. December; 35(21): 7188–7196.
- ↑ DeSantis, T. Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E. L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G. L. (2006). "Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB". Appl Environ Microbiol. 72: 5069–72. doi:10.1128/aem.03006-05.
- ↑ McDonald, D; Price, MN; Goodrich, J; Nawrocki, EP; DeSantis, TZ; Probst, A; Andersen, GL; Knight, R; Hugenholtz, P (2011). "An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea". ISME. 6: 610–618. doi:10.1038/ismej.2011.139.
External links
- University of Washington Laboratory Medicine: Molecular Diagnosis | Bacterial Sequencing
- The Ribosomal Database Project
- Ribosomes and Ribosomal RNA: (rRNA)
- SILVA rRNA database
- Greengenes: 16S rDNA data and tools
- EzBioCloud