Autophagy-related protein 101
| ATG101 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||
| Identifiers | |||||||||||||||||
| Aliases | ATG101, C12orf44, autophagy related 101 | ||||||||||||||||
| External IDs | MGI: 1915368 HomoloGene: 11072 GeneCards: ATG101 | ||||||||||||||||
| |||||||||||||||||
| Orthologs | |||||||||||||||||
| Species | Human | Mouse | |||||||||||||||
| Entrez | |||||||||||||||||
| Ensembl | |||||||||||||||||
| UniProt | |||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||
| RefSeq (protein) | |||||||||||||||||
| Location (UCSC) | Chr 12: 52.07 – 52.08 Mb | Chr 15: 101.28 – 101.29 Mb | |||||||||||||||
| PubMed search | [1] | [2] | |||||||||||||||
| Wikidata | |||||||||||||||||
| View/Edit Human | View/Edit Mouse |
Autophagy-related protein 101 also known as ATG101 is a protein that in humans is encoded by the C12orf44 gene (chromosome 12 open reading frame 44).[3]
Autophagy is the process of forming a vacuole around proteins and nucleic acids that are to be broken down in lysosomes. The transcribed mRNA sequence of C12orf44 is 1287 base pairs, and following translation the sequence is 218 amino acids in length. The ATG101 protein is localized in the cytoplasm, but can possibly also be found bound to a structure known as a phagophore, involved in autophagy. The gene is highly conserved among mammals, as well as showing conservation among most eukaryotes. It is thought to directly interact with ATG13 in the ULK1 complex, which may be important for activating phagophores.
Function
ATG101 is a newly found important autophagy related protein, among many other proteins responsible for autophagy, a process well conserved among most eukaryotic organisms. However, ATG101 is not homologous to any of the other ATG proteins. ATG101 interacts with essential autophagy protein ATG13 in mammals, which is a ULK1 interacting protein.[4] ULK1 (unc-51-like kinase 1) is thought to be important to the activation of macroautophagy in mammals. ATG101 is suggested to protect ATG13 from proteasomal degradation, therefore stabilizing levels of ATG13 found in cells and regulating levels of macroautophagy.[5] According to published papers, ATG101 is said to localize to the isolation membrane, also known as a phagophore. This attachment to the phagophore could possibly explain the palmitoylation site found on the third AA, which is a cysteine. Phagophores are responsible for recognizing and surrounding materials that are to be degraded in the lysosome, by forming a membrane that completely surrounds the materials to be degraded.
Protein properties
| Protein of unknown function DUF1649 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | DUF1649 | ||||||||
| Pfam | PF07855 | ||||||||
| InterPro | IPR012445 | ||||||||
| |||||||||

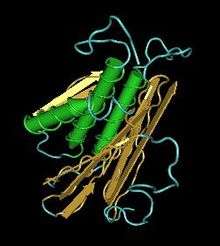
The ATG101 protein contains an domain DUF1649 (domain of unknown function 1649).
ATG101 is highly conserved among most vertebrates, including animals such as mice, whose common ancestor with humans was thought to have diverged between 65 and 85 million years ago. This may be evidence that ATG101 is significant in a way that it must remain conserved structurally in order to retain function. Only 2 missense SNPs are known in the DNA sequence for ATG101, showing that no other versions of this protein exist, also suggesting its structural importance.
In this conceptual translation, the palmitoylation site on the third amino acid can be seen, as well as a possible phosphorylation site shown surrounded by brackets. The amino acids are aligned above the coding region of the mRNA, which is also numbered on the sides. In ATG101, it is interesting to note that this protein is valine-rich.
Gene expression
No concrete data is present establishing ATG101 in any particular area. By looking at multiple expression sets in both humans and mice, it has been hypothesized to be ubiquitously expressed in tissues.
Gene neighborhood
- GRASP GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein[6] -
Codes for a general receptor protein that is relatively unknown, located on the same strand as C12orf44 as seen below, spans 8924 bp.
- NR4A1 (nuclear receptor subfamily 4, group A, member 1) -
This gene encodes nerve growth factor IB that is a member of the steroid-thyroid hormone-retinoid receptor superfamily. Expression is induced by phytohemagglutinin in human lymphocytes and by serum stimulation of arrested fibroblasts. The encoded protein acts as a nuclear transcription factor. Translocation of the protein from the nucleus to mitochondria induces apoptosis. Location is on the same strand as C12orf44, spans 8097 bp.
- OR7E47P (olfactory receptor, family 7, subfamily E, member 47 pseudogene) -
Olfactory receptors interact with odorant molecules in the nose, to initiate a neuronal response that triggers the perception of a smell. The olfactory receptor proteins are members of a large family of G-protein-coupled receptors (GPCR) arising from single coding-exon genes. Olfactory receptors share a 7-transmembrane domain structure with many neurotransmitter and hormone receptors and are responsible for the recognition and G protein-mediated transduction of odorant signals. Located after C12orf44 on the chromosome, it lies on the same strand and is 946 bp in length.
- KRT80 (keratin 80) -
This gene encodes an epithelial keratin that is weakly expressed in the tongue. It is located on the opposite strand to C12orf44 and is 23,005 bp long.
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: chromosome 12 open reading frame 44".
- ↑ Mercer CA, Kaliappan A, Dennis PB (July 2009). "A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy". Autophagy. 5 (5): 649–62. doi:10.4161/auto.5.5.8249. PMID 19287211.
- ↑ Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N (October 2009). "Atg101, a novel mammalian autophagy protein interacting with Atg13". Autophagy. 5 (7): 973–9. doi:10.4161/auto.5.7.9296. PMID 19597335.
- ↑ NM_001098673.1. The NCBI handbook [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2002 Oct. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books