HIST1H4A
| View/Edit Human | View/Edit Mouse |
Histone H4 is a protein that in humans is encoded by the HIST1H4A gene.[3][4][5]
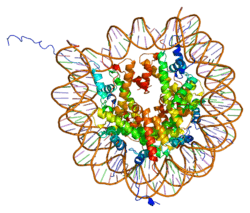
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6.[5]
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D (Apr 1997). "Human histone gene organization: nonregular arrangement within a large cluster". Genomics. 40 (2): 314–22. doi:10.1006/geno.1996.4592. PMID 9119399.
- ↑ Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ (Oct 2002). "The human and mouse replication-dependent histone genes". Genomics. 80 (5): 487–98. doi:10.1016/S0888-7543(02)96850-3. PMID 12408966.
- 1 2 "Entrez Gene: HIST1H4A histone cluster 1, H4a".
Further reading
- Pauli U, Chrysogelos S, Stein G, et al. (1987). "Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene.". Science. 236 (4806): 1308–11. doi:10.1126/science.3035717. PMID 3035717.
- Albig W, Doenecke D (1998). "The human histone gene cluster at the D6S105 locus.". Hum. Genet. 101 (3): 284–94. doi:10.1007/s004390050630. PMID 9439656.
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter.". Mol. Cell. Biol. 18 (5): 2535–44. doi:10.1128/mcb.18.5.2535. PMC 110633
 . PMID 9566873.
. PMID 9566873. - Lorain S, Quivy JP, Monier-Gavelle F, et al. (1998). "Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA.". Mol. Cell. Biol. 18 (9): 5546–56. PMC 109139
 . PMID 9710638.
. PMID 9710638. - Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones.". Virology. 277 (2): 278–95. doi:10.1006/viro.2000.0593. PMID 11080476.
- Freire J, Covelo G, Sarandeses C, et al. (2001). "Identification of nuclear-import and cell-cycle regulatory proteins that bind to prothymosin alpha.". Biochem. Cell Biol. 79 (2): 123–31. doi:10.1139/bcb-79-2-123. PMID 11310559.
- Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA.". Virology. 289 (2): 312–26. doi:10.1006/viro.2001.1129. PMID 11689053.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241
 . PMID 12477932.
. PMID 12477932. - Yoon HG, Chan DW, Huang ZQ, et al. (2003). "Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1.". EMBO J. 22 (6): 1336–46. doi:10.1093/emboj/cdg120. PMC 151047
 . PMID 12628926.
. PMID 12628926. - Lusic M, Marcello A, Cereseto A, Giacca M (2004). "Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter.". EMBO J. 22 (24): 6550–61. doi:10.1093/emboj/cdg631. PMC 291826
 . PMID 14657027.
. PMID 14657027. - Phee H, Abraham RT, Weiss A (2005). "Dynamic recruitment of PAK1 to the immunological synapse is mediated by PIX independently of SLP-76 and Vav1.". Nat. Immunol. 6 (6): 608–17. doi:10.1038/ni1199. PMID 15864311.