Restriction map
A restriction map is a map of known restriction sites within a sequence of DNA. Restriction mapping requires the use of restriction enzymes. In molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by transduction.[1]
One approach in constructing a restriction map of a DNA molecule is to sequence the whole molecule and to run the sequence through a computer program that will find the recognition sites that are present for every restriction enzyme known.
Before sequencing was automated, it would have been prohibitively expensive to sequence an entire DNA strand. To find the relative positions of restriction sites on a plasmid, a technique involving single and double restriction digests is used. Based on the sizes of the resultant DNA fragments the positions of the sites can be inferred. Restriction mapping is a very useful technique when used for determining the orientation of an insert in a cloning vector, by mapping the position of an off-center restriction site in the insert.[2]
Method
The experimental procedure first requires an aliquot of purified plasmid DNA (see appendix) for each digest to be run. Digestion is then performed with each enzyme(s) chosen. The resulting samples are subsequently run on an electrophoresis gel, typically on agarose gel.
The first step following the completion of electrophoresis is to add up the sizes of the fragments in each lane. The sum of the individual fragments should equal the size of the original fragment, and each digest's fragments should also sum up to be the same size as each other. If fragment sizes do not properly add up, there are two likely problems. In one case, some of the smaller fragments may have run off the end of the gel. This frequently occurs if the gel is run too long. A second possible source of error is that the gel was not dense enough and therefore was unable to resolve fragments close in size. This leads to a lack of separation of fragments which were close in size. If all of the digests produce fragments that add up one may infer the position of the REN (restriction endonuclease) sites by placing them in spots on the original DNA fragment that would satisfy the fragment sizes produced by all three digests.
See also restriction enzymes for more detail about the enzymes exploited in this technique.
Example
For example the most common application of restriction mapping is presented: Determining the orientation of a cloned insert. This method requires that restriction maps of the cloning vector and the insert are already available.
If you know of a restriction site placed towards one end of the insert you can determine the orientation by observing the size of the fragments in the gel. Often the orientation of inserts is important and this technique is used to screen for the correct orientation.
In this example the orientation of an insert cloned with EcoRI will be found.
Digests
Resultant Fragments: approximate sizes
- 1: 3 kb, 5 kb
- 2: 2 kb, 6 kb
- 3: 2 kb, 1 kb, 5 kb,
Hypothetical Multiple Cloning Site of Vector
5'-----HindIII-EcoRI----3'
Discussion
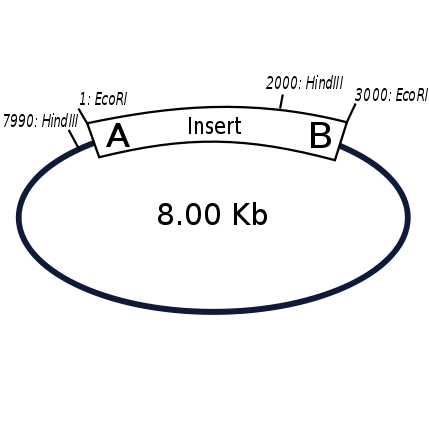
The EcoRI digest excises the insert yielding fragments of 3 kb and 5 kb. These are the sizes of the insert and vector backbone respectively. This is expected since the size of both the insert and vector are known beforehand. The presence of the insert is confirmed.
There is a known HindIII site off-center in the 3 kb insert. It is 2 kb away from one end (end A), and 1 kb away from the other end (end B). The HindIII digest of your clone yields fragments of 2 kb and 6 kb. The 2 kb fragment is exclusively the insert sequence and the 6 kb fragment is 1 kb of insert sequence attached to 5 kb of vector sequence. This means that the insert was cloned in an A to B orientation as opposed to B to A which would yield fragments of 7 kb and 1kb.
resultant map
Appendix: Related Methods
Rapid Denaturation and Renaturation of a crude DNA preparation by alkaline lysis of the cells and subsequent neutralization
In this technique the cells are lysed in alkaline conditions. The DNA in the mixture is denatured (strands separated) by disrupting the hydrogen bonds between the two strands. The large genomic DNA is subject to tangling and staying denatured when the pH is lowered during the neutralization. In other words, the strands come back together in a disordered fashion, basepairing randomly. The circular supercoiled plasmids' strands will stay relatively closely aligned and will renature correctly. Therefore, the genomic DNA will form an insoluble aggregate and the supercoiled plasmids will be left in solution. This can be followed by phenol extraction to remove proteins and other molecules. Then the DNA can be subjected to ethanol precipitation to concentrate the sample.
See also
- Vector NTI - bioinformatics software used among other things to predict restriction sites on a DNA vector
- RFLP- method used to differentiate exceedingly similar genomes, among other things.
References
- ↑ Bitner, R; Kuempel, Peter (February 1982). "P1 Transduction Mapping of the trg Locus in rac+ and rac Strains of Escherichia coli K-12" (PDF). Journal of Bacteriology. 149 (2): 529–533.
- ↑ Dale, J; von Schantz, M; Greenspan, D (2003). From Genes to Genomes. West Sussex: John Wiley & Sons Ltd.