Zetaproteobacteria
| Zetaproteobacteria | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Zetaproteobacteria |
| Species | |
The class Zetaproteobacteria is the sixth and most recently described class of the Proteobacteria.[1] Zetaproteobacteria can also refer to the group of organisms assigned to this class. The Zetaproteobacteria are represented by a single described species, Mariprofundus ferrooxydans,[2] which is an iron-oxidizing neutrophilic chemolithoautotroph originally isolated from Loihi Seamount in 1996 (post-eruption).[1][3] Molecular cloning techniques focusing on the small subunit ribosomal RNA gene have also been used to identify a more diverse majority of the Zetaproteobacteria that have as yet been unculturable.[4]
Regardless of culturing status, the Zetaproteobacteria show up worldwide in estuarine and marine habitats associated with opposing steep redox gradients of reduced (ferrous) iron and oxygen, either as a minor detectable component or as the dominant member of the microbial community.[5][6][7][8][9][10] Zetaproteobacteria have been most commonly found at deep-sea hydrothermal vents,[4] though recent discovery of members of this class in near-shore environments has led to the reevaluation of Zetaproteobacteria distribution and significance.[11][12][13]

Significance
The Zetaproteobacteria are distributed worldwide in deep sea and near shore environments at oxic/anoxic interfaces. With this wide distribution, the Zetaproteobacteria have the potential to play a substantial role in biogeochemical cycling, both past and present. Ecologically, the Zetaproteobacteria play a major role in the engineering of their own environment through the use of the controlled deposition of mineralized iron oxides, also directly affecting the environment of other members of the microbial community.
Prevalence of the Zetaproteobacteria in near-shore metal (e.g. steel) coupon biocorrosion experiments highlights the impact of these marine iron oxidizers on expensive problems such as the rusting of ship hulls, metal pilings, and pipelines.[11][14][15]
Discovery
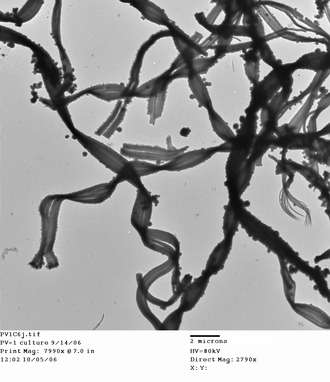
The Zetaproteobacteria were first discovered in 1991 by Craig Moyer, Fred Dobbs, and David Karl as a single rare clone in a mesophilic, or moderate temperature, hydrothermal vent field known as Pele's Vents at Loihi Seamount, Hawaii. This particular vent was dominated by sulfur-oxidizing Epsilonproteobacteria. With no close relatives known at the time, the clone was initially labeled as Gammaproteobacteria.[16]
Subsequent isolation of two strains of M. ferrooxydans, PV-1 and JV-1,[3] along with the increasing realization that a phylogenetically distinct group of Proteobacteria (the Zetaproteobacteria) could be found globally as dominant members of bacterial communities led to the suggestion for the creation of this new class of the Proteobacteria.
Cultivation
Neutrophilic microaerophilic Fe-oxidizing bacteria are typically cultivated using an agarose-stabilized or liquid culture with an FeS or FeCO3 plug. The headspace of the culture tube is then purged with air or a low concentration of oxygen (often 1% or less O2). Fe-oxidizers have also successfully been cultivated in liquid culture with FeCl2 as the Fe source. These cultivation techniques follow those found in Emerson and Floyd (2005).[17]
Recently, researchers have been able to culture the Zetaproteobacteria using graphite electrodes at a fixed voltage.[18] Researchers have also aimed to improve cultivation techniques using a high-biomass batch culturing technique.[19]
Morphology
One of the most distinctive ways of identifying circumneutral iron oxidizing bacteria visually is by identifying the structure of the mineralized iron oxyhydroxide product created during iron oxidation.[3][20] Oxidized, or ferric iron is insoluble at circumneutral pH, thus the microbe must have a way of dealing with the mineralized "waste" product. It is thought that one method to accomplish this is to control the deposition of oxidized iron.[21][22][23] Some of the most common morphotypes include: amorphous particulate oxides, twisted or helical stalks (figure),[21] sheaths,[24] and y-shaped irregular filaments.
These morphologies exist both in freshwater and marine iron habitats, though common freshwater iron-oxidizing bacteria such as Gallionella sp. (twisted stalk) and Leptothrix ochracea (sheath) have only extremely rarely been found in the deep sea (not significant abundance). One currently published morphotype that has been partially resolved is the twisted stalk, which is commonly formed by M. ferrooxydans. This bacteria is a gram negative kidney-bean-shaped cell that deposits iron oxides on the concave side of the cell, forming twisted stalks as it moves through its environment.[21][22]

Another common Zetaproteobacteria morphotype is the sheath structure, which has yet to be isolated, but has been identified with Fluorescence in situ hybridization (FISH).[24]
Iron oxidation morphotypes can be preserved and have been detected in ancient hydrothermal deposits preserved in the rock record.[25][26][27][28][29] Some current work is focused on how the Zetaproteobacteria form their individual biominerals in the modern environment so that scientists can better interpret Fe biominerals found in the rock record.[30][31][32]
Ecology
.png)
Biodiversity
An operational taxonomic unit, or an OTU, allows a microbiologist to define a bacterial taxa using defined similarity bins based on a gene of interest. In microbial ecology, the small subunit ribosomal RNA gene is generally used at a cut off of 97% similarity to define an OTU. In the most basic sense, the OTU represents a bacterial species.
For the Zetaproteobacteria, 28 OTUs have been defined.[4] Of interest were the two globally distributed OTUs that dominated the phylogenetic tree, two OTUs that seemed to originate in the deep subsurface,[10] and several endemic OTUs, along with the relatively limited detection of the isolated Zetaproteobacteria representative.
Classification
Zetaproteobacteria OTUs can now be classified according to the naming scheme used in McAllister et al. (2011).[4] The program ZetaHunter uses closed reference binning to identify sequences closely related to the established OTUs in addition to identifying novel Zetaproteobacteria OTUs. ZetaHunter's feature list continues to grow, but includes: 1) stable OTU binning, 2) sample comparison, 3) database and mask management options, 4) multi-threaded processing, 5) chimera checking, 6) checks for non-database-related sequences, and 7) OTU network maps. The ZetaHunter software can be downloaded at: https://github.com/mooreryan/ZetaHunter
Habitats
- Deep-sea hydrothermal vents associated with:
- Altered deep-sea basalts[49]
- Levantine Basin and continental margin[50]
- Antarctica continental shelf sediment[51]
- Brine/seawater interface[52]
- Stratified Chesapeake Bay estuary[43]
- Intertidal mixing zone of a beach aquifer[13][43]
- Salt marsh sediment[11][53]
- Oxygenated worm burrows or bioturbated beach sands[13][54][55]
- Near-shore metal biocorrosion experiments[11][14]
- Tsunami impacted soils[56]
- Mangrove soils[57]
- Deep subsurface CO2-rich springs[58][59]
- Subsurface flow reactor in the Äspö Hard Rock Laboratory[60]
- Rimicaris exoculata (shrimp) gut at the MAR[61][62]
Ecological Niche
All of the habitats where Zetaproteobacteria have been found have (at least) two things in common: 1) they all provide an interface of steep redox gradients of oxygen and iron.[63] & 2) they are marine or brackish.[44]
Reduced hydrothermal fluids, for instance, exiting from vents in the deep-sea carry with them high concentrations of ferrous iron and other reduced chemical species, creating a gradient upward through a microbial mat of high to low ferrous iron. Similarly, oxygen from the overlying seawater diffuses into the microbial mat resulting in a downward gradient of high to low oxygen. Zetaproteobacteria are thought to live at the interface, where there is enough oxygen for use as an electron acceptor without there being too much oxygen for the organism to compete with the increased rate of chemical oxidation, and where there is enough ferrous iron for growth.[20][63]
Iron oxidation is not always energetically favorable. Reference[39] discusses favorable conditions for iron oxidation in habitats that otherwise may have been thought to be dominated by the more energy yielding metabolisms of hydrogen or sulfur oxidation.
Note: Iron is not the only reduced chemical species accociated with these redox gradient environments. It is likely that Zetaproteobacteria are not all iron oxidizers.
Metabolism
Iron oxidation pathways in both acidophilic and circumneutral freshwater iron oxidation habitats, such as acid mine drainage or groundwater iron seeps, respectively, are better understood than marine circumneutral iron oxidation.
In recent years, researchers have made progress in suggesting possibilities for how the Zetaproteobacteria oxidize iron, primarily through comparative genomics. With this technique, genomes from organisms with similar function, for example the freshwater Fe-oxidizing Betaproteobacteria and the marine Fe-oxidizing Zetaproteobacteria, are compared to find genes that may be required for this function. Identifying the iron oxidation pathway in the Zetaproteobacteria began with the publication of the first described cultured representative, M. ferrooxydans strain PV-1. In this genome, the gene neighborhood of a molybdopterin oxidoreductase protein was identified as a place to start looking at candidate iron oxidation pathway genes.[64] In a follow up analysis of a metagenomic sample, Singer et al. (2013) concluded that this molybdopterin oxidoreductase gene cassette was likely involved in Fe oxidation.[65] Comparative analysis of several single cell genomes, however, suggested an alternative conserved gene cassette with several cytochrome c and cytochrome oxidase genes to be involved in Fe oxidation.[66] For further reading on Fe oxidation pathways see reference.[67]
The phylogenetic distance between the Zetaproteobacteria and the Fe-oxidizing freshwater Betaproteobacteria suggests that Fe oxidation and the produced biominerals are the result of convergent evolution.[24] Comparative genomics has been able to identify several genes that are shared between the two clades, however, suggesting that the trait of Fe oxidation could have been horizontally transferred, possibly virally-mediated.[68][69]
Fe mats associated with the Zetaproteobacteria, in addition to oxidizing Fe have been found to have the genetic potential for denitrification, arsenic detoxification, Calvin-Benson-Bassham (CBB) cycle, and reductive tricarboxylic acid (rTCA) cycles. Novel primers have been designed to detect these genes in environmental samples.[70]
It is difficult at this point to speculate on the metabolism of the entire class of Zetaproteobacteria (with at least 28 different OTUs/species) with the limited sample size.
Suggested Reviews
- Emerson et al., 2010. Iron-oxidizing bacteria: an environmental and genomic perspective.[20]
- Hedrich et al., 2011. The iron-oxidizing proteobacteria.[71]
- Ilbert and Bonnefoy, 2013. Insights into the evolution of the iron oxidation pathways.[67]
- Kato, 2015. Ecophysiology of neutrophilic iron-oxidizing microorganisms and its significance in global biogeochemical cycling.[72]
- Ishibashi et al. eds, 2015. Subseafloor Biosphere Linked to Hydrothermal Systems.[73]
- Melton et al., 2014. The interplay of microbially-mediated and abiotic reactions in the biogeochemical Fe cycle.[74]
See also
References
- 1 2 3 Emerson, D.; Rentz, J. A.; Lilburn, T. G.; Davis, R. E.; Aldrich, H.; Chan, C.; Moyer, C. L. (2007). Reysenbach, Anna-Louise, ed. "A Novel Lineage of Proteobacteria Involved in Formation of Marine Fe-Oxidizing Microbial Mat Communities". PLoS ONE. 2 (8): e667. Bibcode:2007PLoSO...2..667E. doi:10.1371/journal.pone.0000667. PMC 1930151
 . PMID 17668050.
. PMID 17668050. - ↑ mariprofundus entry in LPSN [Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.]
- 1 2 3 Emerson, D.; Moyer, C. L. (2002). "Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition". Applied and Environmental Microbiology. 68 (6): 3085–3093. doi:10.1128/AEM.68.6.3085-3093.2002. PMC 123976
 . PMID 12039770.
. PMID 12039770. - 1 2 3 4 5 McAllister, S. M.; Davis, R. E.; McBeth, J. M.; Tebo, B. M.; Emerson, D.; Moyer, C. L. (2011). "Biodiversity and Emerging Biogeography of the Neutrophilic Iron-Oxidizing Zetaproteobacteria". Applied and Environmental Microbiology. 77 (15): 5445–5457. doi:10.1128/AEM.00533-11. PMC 3147450
 . PMID 21666021.
. PMID 21666021. - 1 2 Schauer, R.; Røy, H.; Augustin, N.; Gennerich, H. H.; Peters, M.; Wenzhoefer, F.; Amann, R.; Meyerdierks, A. (2011). "Bacterial sulfur cycling shapes microbial communities in surface sediments of an ultramafic hydrothermal vent field". Environmental Microbiology. 13 (10): 2633–2648. doi:10.1111/j.1462-2920.2011.02530.x. PMID 21895907.
- 1 2 Hodges, T. W.; Olson, J. B. (2008). "Molecular Comparison of Bacterial Communities within Iron-Containing Flocculent Mats Associated with Submarine Volcanoes along the Kermadec Arc". Applied and Environmental Microbiology. 75 (6): 1650–1657. doi:10.1128/AEM.01835-08. PMC 2655482
 . PMID 19114513.
. PMID 19114513. - 1 2 Davis, R. E.; Stakes, D. S.; Wheat, C. G.; Moyer, C. L. (2009). "Bacterial Variability within an Iron-Silica-Manganese-rich Hydrothermal Mound Located Off-axis at the Cleft Segment, Juan de Fuca Ridge". Geomicrobiology Journal. 26 (8): 570–580. doi:10.1080/01490450902889080.
- 1 2 Forget, N. L.; Murdock, S. A.; Juniper, S. K. (2010). "Bacterial diversity in Fe-rich hydrothermal sediments at two South Tonga Arc submarine volcanoes". Geobiology. 8 (5): 417–432. doi:10.1111/j.1472-4669.2010.00247.x. PMID 20533949.
- 1 2 Handley, K. M.; Boothman, C.; Mills, R. A.; Pancost, R. D.; Lloyd, J. R. (2010). "Functional diversity of bacteria in a ferruginous hydrothermal sediment". The ISME Journal. 4 (9): 1193–1205. doi:10.1038/ismej.2010.38. PMID 20410934.
- 1 2 3 Kato, S.; Yanagawa, K.; Sunamura, M.; Takano, Y.; Ishibashi, J. I.; Kakegawa, T.; Utsumi, M.; Yamanaka, T.; Toki, T.; Noguchi, T.; Kobayashi, K.; Moroi, A.; Kimura, H.; Kawarabayasi, Y.; Marumo, K.; Urabe, T.; Yamagishi, A. (2009). "Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough". Environmental Microbiology. 11 (12): 3210–3222. doi:10.1111/j.1462-2920.2009.02031.x. PMID 19691504.
- 1 2 3 4 McBeth, J. M.; Little, B. J.; Ray, R. I.; Farrar, K. M.; Emerson, D. (2010). "Neutrophilic Iron-Oxidizing "Zetaproteobacteria" and Mild Steel Corrosion in Nearshore Marine Environments". Applied and Environmental Microbiology. 77 (4): 1405–1412. doi:10.1128/AEM.02095-10. PMC 3067224
 . PMID 21131509.
. PMID 21131509. - ↑ McBeth, J. M.; Fleming, E. J.; Emerson, D. (2013). "The transition from freshwater to marine iron-oxidizing bacterial lineages along a salinity gradient on the Sheepscot River, Maine, USA". Environmental Microbiology Reports. 5 (3): 453–63. doi:10.1111/1758-2229.12033. PMID 23754725.
- 1 2 3 McAllister, S. M.; Barnett, J. M.; Heiss, J. W.; Findlay, A. J.; MacDonald, D. J.; Dow, C. L.; Luther, G. W.; Michael, H. A.; Chan, C. S. (2015). "Dynamic hydrologic and biogeochemical processes drive microbially enhanced iron and sulfur cycling within the intertidal mixing zone of a beach aquifer". Limnology and Oceanography. 60: 329–345. doi:10.1002/lno.10029.
- 1 2 Dang, H.; Chen, R.; Wang, L.; Shao, S.; Dai, L.; Ye, Y.; Guo, L.; Huang, G.; Klotz, M. G. (2011). "Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters". Environmental Microbiology. 13 (11): 3059–3074. doi:10.1111/j.1462-2920.2011.02583.x. PMID 21951343.
- ↑ Lee, J. S.; McBeth, J. M.; Ray, R. I.; Little, B. J.; Emerson, D. (2013). "Iron cycling at corroding carbon steel surfaces". Biofouling. 29 (10): 1243–1252. doi:10.1080/08927014.2013.836184.
- ↑ Moyer, C. L.; Dobbs, F. C.; Karl, D. M. (1995). "Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii". Applied and Environmental Microbiology. 61 (4): 1555–1562. PMC 167411
 . PMID 7538279.
. PMID 7538279. - ↑ Emerson, D.; Merrill Floyd, M. (2005). "Enrichment and Isolation of Iron‐Oxidizing Bacteria at Neutral pH". Environmental Microbiology. Methods in Enzymology. 397. p. 112. doi:10.1016/S0076-6879(05)97006-7. ISBN 9780121828028.
- ↑ Summers, Z. M.; Gralnick, J. A.; Bond, D. R. (2013). "Cultivation of an Obligate Fe(II)-Oxidizing Lithoautotrophic Bacterium Using Electrodes". MBio. 4 (1): e00420–e00412. doi:10.1128/mBio.00420-12. PMC 3560526
 . PMID 23362318.
. PMID 23362318. - ↑ Barco, R. A.; Edwards, K. J. (2014). "Interactions of proteins with biogenic iron oxyhydroxides and a new culturing technique to increase biomass yields of neutrophilic, iron-oxidizing bacteria". Frontiers in Microbiology. 5. doi:10.3389/fmicb.2014.00259.
- 1 2 3 Emerson, D.; Fleming, E. J.; McBeth, J. M. (2010). "Iron-Oxidizing Bacteria: An Environmental and Genomic Perspective". Annual Review of Microbiology. 64: 561–583. doi:10.1146/annurev.micro.112408.134208. PMID 20565252.
- 1 2 3 Chan, C. S.; Fakra, S. C.; Emerson, D.; Fleming, E. J.; Edwards, K. J. (2010). "Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: Implications for biosignature formation". The ISME Journal. 5 (4): 717–727. doi:10.1038/ismej.2010.173. PMC 3105749
 . PMID 21107443.
. PMID 21107443. - 1 2 Comolli, L. R.; Luef, B.; Chan, C. S. (2011). "High-resolution 2D and 3D cryo-TEM reveals structural adaptations of two stalk-forming bacteria to an Fe-oxidizing lifestyle". Environmental Microbiology. 13 (11): 2915–2929. doi:10.1111/j.1462-2920.2011.02567.x. PMID 21895918.
- ↑ Saini, G.; Chan, C. S. (2013). "Near-neutral surface charge and hydrophilicity prevent mineral encrustation of Fe-oxidizing micro-organisms". Geobiology. 11 (2): 191–200. doi:10.1111/gbi.12021. PMID 23279435.
- 1 2 3 Fleming, E. J.; Davis, R. E.; McAllister, S. M.; Chan, C. S.; Moyer, C. L.; Tebo, B. M.; Emerson, D. (2013). "Hidden in plain sight: Discovery of sheath-forming, iron-oxidizing Zetaproteobacteriaat Loihi Seamount, Hawaii, USA". FEMS Microbiology Ecology. 85: 116–127. doi:10.1111/1574-6941.12104.
- ↑ Juniper, S. Kim; Yves Fouquet (1988). "Filamentous iron-silica deposits from modern and ancient hydrothermal sites". Canadian Mineralogist. 26: 859–869.
- ↑ Hofmann, B. A.; Farmer, J. D.; von Blanckenburg, F.; Fallick, A. E. (2008). "Subsurface Filamentous Fabrics: An Evaluation of Origins Based on Morphological and Geochemical Criteria, with Implications for Exopaleontology". Astrobiology. 8 (1): 87–117. doi:10.1089/ast.2007.0130. PMID 18241094.
- ↑ Little, C. T. S.; Glynn, S. E. J.; Mills, R. A. (2004). "Four-Hundred-and-Ninety-Million-Year Record of Bacteriogenic Iron Oxide Precipitation at Sea-Floor Hydrothermal Vents". Geomicrobiology Journal. 21 (6): 415–429. doi:10.1080/01490450490485845.
- ↑ Sun, Z.; Li, J.; Huang, W.; Dong, H.; Little, C. T. S.; Li, J. (2015). "Generation of hydrothermal Fe-Si oxyhydroxide deposit on the Southwest Indian Ridge and its implication for the origin of ancient banded iron formations". Journal of Geophysical Research: Biogeosciences. 120: 187–203. doi:10.1002/2014JG002764.
- ↑ Krepski, S. T.; Emerson, D.; Hredzak-Showalter, P. L.; Luther, G. W.; Chan, C. S. (2013). "Morphology of biogenic iron oxides records microbial physiology and environmental conditions: Toward interpreting iron microfossils". Geobiology. 11 (5): 457–71. doi:10.1111/gbi.12043. PMID 23790206.
- ↑ Chan, C. S.; Fakra, S. C.; Edwards, D. C.; Emerson, D.; Banfield, J. F. (2009). "Iron oxyhydroxide mineralization on microbial extracellular polysaccharides". Geochimica et Cosmochimica Acta. 73 (13): 3807–3818. Bibcode:2009GeCoA..73.3807C. doi:10.1016/j.gca.2009.02.036.
- ↑ Bennett, S. A.; Toner, B. M.; Barco, R.; Edwards, K. J. (2014). "Carbon adsorption onto Fe oxyhydroxide stalks produced by a lithotrophic iron-oxidizing bacteria". Geobiology. 12 (2): 146–156. doi:10.1111/gbi.12074.
- ↑ Rassa, A. C.; McAllister, S. M.; Safran, S. A.; Moyer, C. L. (2009). "Zeta-ProteobacteriaDominate the Colonization and Formation of Microbial Mats in Low-Temperature Hydrothermal Vents at Loihi Seamount, Hawaii". Geomicrobiology Journal. 26 (8): 623–638. doi:10.1080/01490450903263350.
- ↑ Emerson, D.; Moyer, C. (2010). "Microbiology of Seamounts: Common Patterns Observed in Community Structure". Oceanography. 23: 148–163. doi:10.5670/oceanog.2010.67.
- ↑ Sudek, L. A.; Templeton, A. S.; Tebo, B. M.; Staudigel, H. (2009). "Microbial Ecology of Fe (hydr)oxide Mats and Basaltic Rock from Vailulu'u Seamount, American Samoa". Geomicrobiology Journal. 26 (8): 581–596. doi:10.1080/01490450903263400.
- ↑ Edwards, K. J.; Glazer, B. T.; Rouxel, O. J.; Bach, W.; Emerson, D.; Davis, R. E.; Toner, B. M.; Chan, C. S.; Tebo, B. M.; Staudigel, H.; Moyer, C. L. (2011). "Ultra-diffuse hydrothermal venting supports Fe-oxidizing bacteria and massive umber deposition at 5000 m off Hawaii". The ISME Journal. 5 (11): 1748–1758. doi:10.1038/ismej.2011.48. PMC 3197161
 . PMID 21544100.
. PMID 21544100. - ↑ Kato, S.; Kobayashi, C.; Kakegawa, T.; Yamagishi, A. (2009). "Microbial communities in iron-silica-rich microbial mats at deep-sea hydrothermal fields of the Southern Mariana Trough". Environmental Microbiology. 11 (8): 2094–2111. doi:10.1111/j.1462-2920.2009.01930.x. PMID 19397679.
- ↑ Davis, R. E.; Moyer, C. L. (2008). "Extreme spatial and temporal variability of hydrothermal microbial mat communities along the Mariana Island Arc and southern Mariana back-arc system". Journal of Geophysical Research. 113. Bibcode:2008JGRB..113.8S15D. doi:10.1029/2007JB005413.
- 1 2 Kato, S.; Nakamura, K.; Toki, T.; Ishibashi, J. I.; Tsunogai, U.; Hirota, A.; Ohkuma, M.; Yamagishi, A. (2012). "Iron-Based Microbial Ecosystem on and Below the Seafloor: A Case Study of Hydrothermal Fields of the Southern Mariana Trough". Frontiers in Microbiology. 3. doi:10.3389/fmicb.2012.00089.
- ↑ Meyer-Dombard, D. A. R.; Amend, J. P.; Osburn, M. R. (2012). "Microbial diversity and potential for arsenic and iron biogeochemical cycling at an arsenic rich, shallow-sea hydrothermal vent (Tutum Bay, Papua New Guinea)". Chemical Geology. 348: 37–47. doi:10.1016/j.chemgeo.2012.02.024.
- ↑ Li, J.; Zhou, H.; Peng, X.; Wu, Z.; Chen, S.; Fang, J. (2012). "Microbial diversity and biomineralization in low-temperature hydrothermal iron-silica-rich precipitates of the Lau Basin hydrothermal field". FEMS Microbiology Ecology. 81 (1): 205–216. doi:10.1111/j.1574-6941.2012.01367.x. PMID 22443540.
- ↑ Dekov, V. M.; Petersen, S.; Garbe-Schönberg, C. -D.; Kamenov, G. D.; Perner, M.; Kuzmann, E.; Schmidt, M. (2010). "Fe–Si-oxyhydroxide deposits at a slow-spreading centre with thickened oceanic crust: The Lilliput hydrothermal field (9°33′S, Mid-Atlantic Ridge)". Chemical Geology. 278 (3–4): 186–200. doi:10.1016/j.chemgeo.2010.09.012.
- 1 2 3 MacDonald, D. J.; Findlay, A. J.; McAllister, S. M.; Barnett, J. M.; Hredzak-Showalter, P.; Krepski, S. T.; Cone, S. G.; Scott, J.; Bennett, S. K.; Chan, C. S.; Emerson, D.; Luther Iii, G. W. (2014). "Using in situ voltammetry as a tool to identify and characterize habitats of iron-oxidizing bacteria: From fresh water wetlands to hydrothermal vent sites". Environmental Science: Processes & Impacts. 16 (9): 2117. doi:10.1039/c4em00073k.
- 1 2 Scott, J. J.; Breier, J. A.; Luther, G. W.; Emerson, D. (2015). "Microbial Iron Mats at the Mid-Atlantic Ridge and Evidence that Zetaproteobacteria May Be Restricted to Iron-Oxidizing Marine Systems". PLoS ONE. 10 (3): e0119284. doi:10.1371/journal.pone.0119284. PMC 4356598
 . PMID 25760332.
. PMID 25760332. - ↑ Cao, H.; Wang, Y.; Lee, O. O.; Zeng, X.; Shao, Z.; Qian, P. -Y. (2014). "Microbial Sulfur Cycle in Two Hydrothermal Chimneys on the Southwest Indian Ridge". MBio. 5 (1): e00980–e00913. doi:10.1128/mBio.00980-13. PMC 3903282
 . PMID 24473131.
. PMID 24473131. - ↑ Sylvan, J. B.; Toner, B. M.; Edwards, K. J. (2012). "Life and Death of Deep-Sea Vents: Bacterial Diversity and Ecosystem Succession on Inactive Hydrothermal Sulfides". MBio. 3 (1): e00279–e00211. doi:10.1128/mBio.00279-11. PMC 3262234
 . PMID 22275502.
. PMID 22275502. - ↑ Dhillon, A.; Teske, A.; Dillon, J.; Stahl, D. A.; Sogin, M. L. (2003). "Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin". Applied and Environmental Microbiology. 69 (5): 2765–2772. doi:10.1128/AEM.69.5.2765-2772.2003. PMC 154542
 . PMID 12732547.
. PMID 12732547. - ↑ Kato, S.; Ikehata, K.; Shibuya, T.; Urabe, T.; Ohkuma, M.; Yamagishi, A. (2015). "Potential for biogeochemical cycling of sulfur, iron and carbon within massive sulfide deposits below the seafloor". Environmental Microbiology. 17 (5): 1817–35. doi:10.1111/1462-2920.12648. PMID 25330135.
- ↑ Jacobson Meyers, M. E.; Sylvan, J. B.; Edwards, K. J. (2014). "Extracellular Enzyme Activity and Microbial Diversity Measured on Seafloor Exposed Basalts from Loihi Seamount Indicate the Importance of Basalts to Global Biogeochemical Cycling". Applied and Environmental Microbiology. 80 (16): 4854–64. doi:10.1128/AEM.01038-14. PMID 24907315.
- ↑ Rubin-Blum, M.; Antler, G.; Tsadok, R.; Shemesh, E.; Austin, J. A.; Coleman, D. F.; Goodman-Tchernov, B. N.; Ben-Avraham, Z.; Tchernov, D. (2014). "First Evidence for the Presence of Iron Oxidizing Zetaproteobacteria at the Levantine Continental Margins". PLoS ONE. 9 (3): e91456. doi:10.1371/journal.pone.0091456. PMC 3948872
 . PMID 24614177.
. PMID 24614177. - ↑ Bowman, J. P.; McCuaig, R. D. (2003). "Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment". Applied and Environmental Microbiology. 69 (5): 2463–2483. doi:10.1128/AEM.69.5.2463-2483.2003. PMC 154503
 . PMID 12732511.
. PMID 12732511. - ↑ Eder, W.; Jahnke, L. L.; Schmidt, M.; Huber, R. (2001). "Microbial Diversity of the Brine-Seawater Interface of the Kebrit Deep, Red Sea, Studied via 16S rRNA Gene Sequences and Cultivation Methods". Applied and Environmental Microbiology. 67 (7): 3077–3085. doi:10.1128/AEM.67.7.3077-3085.2001. PMC 92984
 . PMID 11425725.
. PMID 11425725. - ↑ Moreau, J. W.; Zierenberg, R. A.; Banfield, J. F. (2010). "Diversity of Dissimilatory Sulfite Reductase Genes (dsrAB) in a Salt Marsh Impacted by Long-Term Acid Mine Drainage". Applied and Environmental Microbiology. 76 (14): 4819–4828. doi:10.1128/AEM.03006-09. PMC 2901737
 . PMID 20472728.
. PMID 20472728. - ↑ Stauffert, M.; Cravo-Laureau, C.; Jézéquel, R.; Barantal, S.; Cuny, P.; Gilbert, F.; Cagnon, C.; Militon, C. C.; Amouroux, D.; Mahdaoui, F.; Bouyssiere, B.; Stora, G.; Merlin, F. O. X.; Duran, R. (2013). "Impact of Oil on Bacterial Community Structure in Bioturbated Sediments". PLoS ONE. 8 (6): e65347. doi:10.1371/journal.pone.0065347.
- ↑ Pischedda, L.; Militon, C. C.; Gilbert, F.; Cuny, P. (2011). "Characterization of specificity of bacterial community structure within the burrow environment of the marine polychaete Hediste (Nereis) diversicolor". Research in Microbiology. 162 (10): 1033–42. doi:10.1016/j.resmic.2011.07.008. PMID 21946148.
- ↑ Asano, R.; Nakai, Y.; Kawada, W.; Shimura, Y.; Inamoto, T.; Fukushima, J. (2013). "Seawater Inundation from the 2011 Tohoku Tsunami Continues to Strongly Affect Soil Bacterial Communities 1 Year Later". Microbial Ecology. 66 (3): 639–46. doi:10.1007/s00248-013-0261-9. PMID 23846833.
- ↑ Thompson, C.; Beys-Da-Silva, W.; Santi, L. L.; Berger, M.; Vainstein, M.; Guima Rães, J.; Vasconcelos, A. T. (2013). "A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves". AMB Express. 3 (1): 65. doi:10.1186/2191-0855-3-65. PMC 3922913
 . PMID 24160319.
. PMID 24160319. - ↑ Emerson, J. B.; Thomas, B. C.; Alvarez, W.; Banfield, J. F. (2015). "Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla". Environmental Microbiology: n/a. doi:10.1111/1462-2920.12817.
- ↑ Colman, D. R.; Garcia, J. R.; Crossey, L. J.; Karlstrom, K.; Jackson-Weaver, O.; Takacs-Vesbach, C. (2014). "An analysis of geothermal and carbonic springs in the western United States sustained by deep fluid inputs". Geobiology. 12: 83–98. doi:10.1111/gbi.12070.
- ↑ Ionescu, D.; Heim, C.; Polerecky, L.; Ramette, A.; Haeusler, S.; Bizic-Ionescu, M.; Thiel, V.; De Beer, D. (2015). "Diversity of Iron Oxidizing and Reducing Bacteria in Flow Reactors in the Äspö Hard Rock Laboratory". Geomicrobiology Journal. 32 (3–4): 207–220. doi:10.1080/01490451.2014.884196.
- ↑ Zbinden, M.; Cambon-Bonavita, M. A. (2003). "Occurrence of Deferribacterales and Entomoplasmatales in the deep-sea Alvinocarid shrimp Rimicaris exoculata gut". FEMS Microbiology Ecology. 46 (1): 23–30. doi:10.1016/S0168-6496(03)00176-4. PMID 19719579.
- ↑ Jan, C.; Petersen, J. M.; Werner, J.; Teeling, H.; Huang, S.; Glöckner, F. O.; Golyshina, O. V.; Dubilier, N.; Golyshin, P. N.; Jebbar, M.; Cambon-Bonavita, M. A. (2014). "The gill chamber epibiosis of deep-sea shrimp Rimicarisexoculata: An in-depth metagenomic investigation and discovery of Zetaproteobacteria". Environmental Microbiology. 16 (9): 2723–38. doi:10.1111/1462-2920.12406. PMID 24447589.
- 1 2 Glazer, B. T.; Rouxel, O. J. (2009). "Redox Speciation and Distribution within Diverse Iron-dominated Microbial Habitats at Loihi Seamount". Geomicrobiology Journal. 26 (8): 606–622. doi:10.1080/01490450903263392.
- ↑ Singer, E.; Emerson, D.; Webb, E. A.; Barco, R. A.; Kuenen, J. G.; Nelson, W. C.; Chan, C. S.; Comolli, L. R.; Ferriera, S.; Johnson, J.; Heidelberg, J. F.; Edwards, K. J. (2011). Khodursky, Arkady B, ed. "Mariprofundus ferrooxydans PV-1 the First Genome of a Marine Fe(II) Oxidizing Zetaproteobacterium". PLoS ONE. 6 (9): e25386. doi:10.1371/journal.pone.0025386. PMC 3179512
 . PMID 21966516.
. PMID 21966516. - ↑ Singer, E.; Heidelberg, J. F.; Dhillon, A.; Edwards, K. J. (2013). "Metagenomic insights into the dominant Fe(II) oxidizing Zetaproteobacteria from an iron mat at Lō´ihi, Hawai´l". Frontiers in Microbiology. 4. doi:10.3389/fmicb.2013.00052.
- ↑ Field, E. K.; Sczyrba, A.; Lyman, A. E.; Harris, C. C.; Woyke, T.; Stepanauskas, R.; Emerson, D. (2014). "Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount". The ISME Journal. 9 (4): 857–70. doi:10.1038/ismej.2014.183. PMID 25303714.
- 1 2 Ilbert, M.; Bonnefoy, V. (2013). "Insight into the evolution of the iron oxidation pathways". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1827 (2): 161–175. doi:10.1016/j.bbabio.2012.10.001.
- ↑ Emerson, D.; Field, E. K.; Chertkov, O.; Davenport, K. W.; Goodwin, L.; Munk, C.; Nolan, M.; Woyke, T. (2013). "Comparative genomics of freshwater Fe-oxidizing bacteria: Implications for physiology, ecology, and systematics". Frontiers in Microbiology. 4. doi:10.3389/fmicb.2013.00254.
- ↑ Altermann, E. (2014). "Invited commentary: Lubricating the rusty wheel, new insights into iron oxidizing bacteria through comparative genomics". Frontiers in Microbiology. 5. doi:10.3389/fmicb.2014.00386.
- ↑ Jesser, K. J.; Fullerton, H.; Hager, K. W.; Moyer, C. L. (2015). "Quantitative PCR Analysis of Functional Genes in Iron-Rich Microbial Mats at an Active Hydrothermal Vent System (Lō'ihi Seamount, Hawai'i)". Applied and Environmental Microbiology. 81 (9): 2976–2984. doi:10.1128/AEM.03608-14.
- ↑ Hedrich, S.; Schlomann, M.; Johnson, D. B. (2011). "The iron-oxidizing proteobacteria". Microbiology. 157 (6): 1551–1564. doi:10.1099/mic.0.045344-0.
- ↑ Kato, Shingo (2015). "Ecophysiology of neutrophilic iron-oxidizing microorganisms and its significance in global biogeochemical cycling". Chikyukagaku (Geochemistry). 49: 1–17. doi:10.14934/chikyukagakuI.1.
- ↑ "Subseafloor Biosphere Linked to Hydrothermal Systems". 2015. doi:10.1007/978-4-431-54865-2. ISBN 978-4-431-54864-5.
- ↑ Melton, E. D.; Swanner, E. D.; Behrens, S.; Schmidt, C.; Kappler, A. (2014). "The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle". Nature Reviews Microbiology. 12 (12): 797–808. doi:10.1038/nrmicro3347. PMID 25329406.