Centrosome cycle
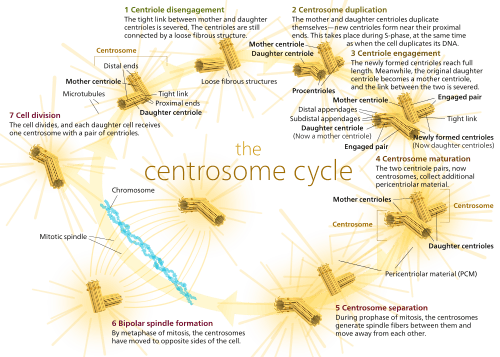
Centrosomes are the major microtubule organizing center (MTOC) in mammalian cells.[2] Failure of centrosome regulation can cause mistakes in chromosome segregation and is associated with aneuploidy. A centrosome is composed of two orthogonal cylindrical proteins, called centrioles, which are surrounded by an electron dense and protein dense amorphous cloud of pericentriolar matrix, often abbreviated as "PCM".[3] The PCM is essential for nucleation and organization of microtubules.[3] The centrosome cycle is important to ensure that daughter cells receive a centrosome after cell division. As the cell cycle progresses, the centrosome undergoes a series of morphological and functional changes. Initiation of the centrosome cycle occurs early in the cell cycle, so that by the time mitosis occurs there are two centrosomes.
The centrosome cycle consists of four phases that are synchronized to cell cycle. These include: centrosome duplication during the G1 phase and S Phase, centrosome maturation in the G2 phase, centrosome separation in the mitotic phase, and centrosome disorientation in the late mitotic phase—G1 phase.
Centriole synthesis
Centrioles are generated in new daughter cells through duplication of preexisting centrioles in the mother cells. Each daughter cell inherits two centrioles (one centrosome) surrounded by pericentriolar material as a result of cell division. The two centrioles are of different ages, though. One centriole originates from the mother cell and the other is replicated from the mother centriole during the cell cycle. It is possible to distinguish between the two preexisting centrioles, because the mother and daughter centriole differ in both shape and function.[4] For example, the mother centriole can nucleate and organize microtubules, whereas the daughter centriole can only nucleate.
First, procentrioles begin to form near each preexisting centriole as the cell moves from the G1 phase to the S phase.[5][6][7] During S and G2 phase of the cell cycle, the procentrioles elongate until they reach the length of the older mother and daughter centrioles(which takes on characteristics of a mother centriole). Once they reach full length, the new centriole and its mother centriole form a diplosome. A diplosome is a rigid complex formed by an orthogonal mother and newly formed centriole (now a daughter centriole) that aids in the processes of mitosis. As mitosis occurs, the distance between mother and daughter centriole increases until, congruent with anaphase, the diplosome breaks down and each centriole is surrounded by its own pericentriolar material.[5]
Centrosome Duplication
Cell Cycle Regulation of Centrosome Duplication
Centrosome duplication is heavily regulated by cell cycle controls. This link between the cell cycle and the centrosome cycle is mediated by cyclin-dependent kinase 2 (Cdk2). There has been ample evidence [8][9][10][11] that Cdk2 is necessary for both DNA replication and centrosome duplication, which are both key events in S phase. It has also been shown [10][12][13] that Cdk2 complexes with both cyclin A and cyclin E and this complex is critical for centrosome duplication. Three Cdk2 substrates have been proposed to be responsible for regulation of centriole duplication. These include: nucleophosmin (NPM/B23), CP110, and MPS1.[3] Nucleophosmin is only found in unreplicated centrosomes and it’s phosphorylation by Cdk2/cyclin E removes NPM from the centrosomes, initiating procentriole formation.[14][15] CP110 is an important centrosomal protein that is phosphorylated by both mitotic and interphase Cdk/cyclin complexes and is thought to influence centrosome duplication in S phase. [19] MPS1 is a protein kinase that is essential to the spindle assembly checkpoint,[16] and may remodel an SAS6-cored intermediate between severed mother and daughter centrioles into a pair of cartwheel protein complexes onto which procentrioles assemble.[17]
Centrosome Maturation
Centrosome maturation is defined as the increase or accumulation of γ-tubulin ring complexes and other PCM proteins at the centrosome.[2] This increase in γ -tubulin allows the mature centrosome to have a greater ability to nucleate microtubules. Phosphorylation is a key regulatory role in centrosome maturation and it is thought that Polo-like kinases (Plks) and Aurora kinases are responsible for this phosphorylation. [21] The phosphorylation of downstream targets of Plks and Aurora A lead to the recruitment of γ –tubulin and other proteins that form PCM around the centrioles. [23]
Centrosome Separation
In early mitosis, several motor proteins drive the separation of centrosomes. With the onset of prophase, the motor protein, dynein, provides the majority of the force required to pull the two centrosomes apart. The separation event actually occurs at the G2/M transition and happens in two steps. First, the connection between the two parental centrioles is destroyed. Second, the centrosomes are separated via microtubules motor proteins.[2]
Centrosome Disorientation
Centrosome disorientation refers to the loss of orthogonality between the mother and daughter centrioles.[2] Once disorientation occurs, the mature centriole begins to move toward the cleave furrow and it was purposed that this movement is a key step in abscission, the terminal phase of cell division.[18]
Disregulation of the Centrosome Cycle
Improper progression through the centrosome cycle can lead to incorrect numbers of centrosomes and aneuploidy, which could eventually lead to cancer. The role of centrosomes in tumor progression is unclear. The misexpression of genes such as, p53, BRCA1, Mdm2, Aurora-A and survivin, causes an increase in the amount of centrosomes present in a cell. However, it is not well understood how these genes influence the centrosome or how the increase number of centrosomes influences tumor progression.[19]
References
- ↑ "Figure 1". Aurora-A: the maker and breaker of spindle poles. Journal of Cell Science. Retrieved 11 December 2012.
- 1 2 3 4 Meraldi, P; Nigg, E.A. (23 April 2002). "The centrosome cycle". FEBS Letters. 521 (1–3): 9–13. doi:10.1016/S0014-5793(02)02865-X. PMID 12067716.
- 1 2 3 Loncarek, J.; Khodjakov, A. (February 2009). "Ab ovo or de novo? Mechanisms of centriole duplication". Mol Cells. 27 (2): 135–142. doi:10.1007/s10059-009-0017-z. PMC 2691869
 . PMID 19277494.
. PMID 19277494. - ↑ Piel, M; Nordberg, J; Euteneuer, U; Bornens, M (Feb 23, 2001). "Centrosome-dependent exit of cytokinesis in animal cells". Science. 291 (5508): 1550–3. doi:10.1126/science.291.5508.1550. PMID 11222861.
- 1 2 Chrétien, D; Buendia, B; Fuller, SD; Karsenti, E (November 1997). "Reconstruction of the centrosome cycle from cryoelectron micrographs". Journal of structural biology. 120 (2): 117–33. doi:10.1006/jsbi.1997.3928. PMID 9417977.
- ↑ Kuriyama, R; Borisy, GG (December 1981). "Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy". The Journal of Cell Biology. 91 (3 Pt 1): 814–21. doi:10.1083/jcb.91.3.814. PMC 2112828
 . PMID 7328123.
. PMID 7328123. - ↑ Vorobjev, IA; Chentsov, YuS (June 1982). "Centrioles in the cell cycle. I. Epithelial cells". The Journal of Cell Biology. 93 (3): 938–49. doi:10.1083/jcb.93.3.938. PMC 2112136
 . PMID 7119006.
. PMID 7119006. - ↑ Hinchcliffe, EH; Li, C; Thompson, EA; Maller, JL; Sluder, G (Feb 5, 1999). "Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts". Science. 283 (5403): 851–4. doi:10.1126/science.283.5403.851. PMID 9933170.
- ↑ Matsumoto, Y; Hayashi, K; Nishida, E (Apr 22, 1999). "Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells". Current Biology. 9 (8): 429–32. doi:10.1016/S0960-9822(99)80191-2. PMID 10226033.
- 1 2 Meraldi, P; Lukas, J; Fry, AM; Bartek, J; Nigg, EA (June 1999). "Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A". Nature Cell Biology. 1 (2): 88–93. doi:10.1038/10054. PMID 10559879.
- ↑ Lacey, KR; Jackson, PK; Stearns, T (Mar 16, 1999). "Cyclin-dependent kinase control of centrosome duplication". Proceedings of the National Academy of Sciences of the United States of America. 96 (6): 2817–22. doi:10.1073/pnas.96.6.2817. PMC 15852
 . PMID 10077594.
. PMID 10077594. - ↑ Hinchcliffe, EH; Sluder, G. (2001b). "Centrosome duplication: Three kinases come up a winner!". Cur Biol. 11 (17): R698–R701. doi:10.1016/S0960-9822(01)00412-2.
- ↑ Matsumoto, Y; Maller, JL (Oct 29, 2004). "A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry". Science. 306 (5697): 885–8. doi:10.1126/science.1103544. PMID 15514162.
- ↑ Okuda, M; Horn, HF; Tarapore, P; Tokuyama, Y; Smulian, AG; Chan, PK; Knudsen, ES; Hofmann, IA; Snyder, JD; Bove, KE; Fukasawa, K (Sep 29, 2000). "Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication". Cell. 103 (1): 127–40. doi:10.1016/S0092-8674(00)00093-3. PMID 11051553.
- ↑ Tokuyama, Y; Horn, HF; Kawamura, K; Tarapore, P; Fukasawa, K (Jun 15, 2001). "Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication". The Journal of Biological Chemistry. 276 (24): 21529–37. doi:10.1074/jbc.M100014200. PMID 11278991.
- ↑ Stucke, VM; Silljé, HH; Arnaud, L; Nigg, EA (Apr 2, 2002). "Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication". The EMBO Journal. 21 (7): 1723–32. doi:10.1093/emboj/21.7.1723. PMC 125937
 . PMID 11927556.
. PMID 11927556. - ↑ Pike, Amanda N; Fisk, Harold N (April 14, 2011). "Centriole assembly and the role of Mps1: defensible or dispensable?". Cell Division. 6: 6–14. doi:10.1186/1747-1028-6-9. PMC 3094359
 . PMID 21492451.
. PMID 21492451. - ↑ Rusan, Nasser M.; Rogers, Gregory C. (1 May 2009). "Centrosome Function: Sometimes Less Is More". Traffic. 10 (5): 472–481. doi:10.1111/j.1600-0854.2009.00880.x. PMID 19192251.
- ↑ Cunha-Ferreira, Inês; Bento, Inês; Bettencourt-Dias, Mónica (1 May 2009). "From Zero to Many: Control of Centriole Number in Development and Disease". Traffic. 10 (5): 482–498. doi:10.1111/j.1600-0854.2009.00905.x. PMID 19416494.