Complement membrane attack complex
.png)
The membrane attack complex (MAC) or terminal complement complex (TCC) is a structure typically formed on the surface of pathogenic bacterial cells as a result of the activation of the host's alternative pathway, classical pathway, or lectin pathway of the complement system, and it is one of the effector proteins of the immune system. The membrane-attack complex (MAC) forms transmembrane channels. These channels disrupt the cell membrane of target cells, leading to cell lysis and death.[1][2]
A number of proteins participate in the assembly of the MAC. Freshly activated C5b binds to C6 to form a C5b-6 complex, then to C7 forming the C5b-6-7 complex. The C5b-6-7 complex binds to C8, which is composed of three chains (alpha, beta, and gamma), thus forming the C5b-6-7-8 complex. C5b-6-7-8 subsequently binds to C9[3][4][5] and acts as a catalyst in the polymerization of C9.
Active MAC has a subunit composition of C5b-C6-C7-C8-C9{n}.
Structure and function
It is composed of a complex of four complement proteins (C5b, C6, C7, and C8) that bind to the outer surface of the plasma membrane, and many copies of a fifth protein (C9) that hook up to one another, forming a ring in the membrane. C6-C9 all contain a common MACPF domain.[6] This region is homologous to cholesterol-dependent cytolysins from Gram-positive bacteria.[7]
The ring structure formed by C9 is a pore in the membrane that allows free diffusion of molecules in and out of the cell. If enough pores form, the cell is no longer able to survive.
If the pre-MAC complexes of C5b-7, C5b-8 or C5b-9 do not insert into a membrane, they can form inactive complexes with Protein S (sC5b-7, sC5b-8 and sC5b-9). These fluid phase complexes do not bind to cell membranes and are ultimately scavenged by clusterin and vitronectin, two regulators of complement.[8]
Initiation: C5-C7
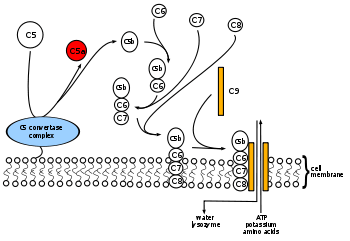
The membrane attack complex is initiated when the complement protein C5 convertase cleaves C5 into C5a and C5b.
Another complement protein, C6, binds to C5b.
The C5bC6 complex is bound by C7.
This junction alters the configuration of the protein molecules exposing a hydrophobic site on C7 that allows the C7 to insert into the phospholipid bilayer of the pathogen.
Polymerization: C8-C9
Similar hydrophobic sites on C8 and C9 molecules are exposed when they bind to the complex, so they can also insert into the bilayer.
C8 is a complex made of the two proteins C8-beta and C8 alpha-gamma.
C8 alpha-gamma has the hydrophobic area that inserts into the bilayer. C8 alpha-gamma induces the polymerization of 10-16 molecules of C9 into a pore-forming structure known as the membrane attack complex.
- It has a hydrophobic external face allowing it to associate with the lipid bilayer.
- It has a hydrophilic internal face to allow the passage of water.
Multiple molecules of C9 can join spontaneously in concentrated solution to form polymers of C9. These polymers can also form a tube-like structure.
Inhibition
CD59 acts to inhibit the complex. This exists on body cells to protect them from MAC. A rare condition, paroxysmal nocturnal haemoglobinuria, results in red cells that lack CD59. These red cells can, therefore, be lysed by MAC.
Pathology
Deficiencies of C5 to C9 components does not lead to generic infections, but only to increased susceptibility to Neisseria spp.,[9] since these bacteria have a thin cell wall and glycocalix.[10]
References
- ↑ Peitsch MC, Tschopp J (1991). "Assembly of macromolecular pores by immune defense systems". Curr. Opin. Cell Biol. 3 (4): 710–716. doi:10.1016/0955-0674(91)90045-Z. PMID 1722985.
- ↑ Reid K. B. M., The complement system, in: B. D. Hames and D. M. Glover (eds.), Molecular Immunology, Oxford: IRL Press, 1988, pp. 189-241.
- ↑ Stanley KK, Marazziti D, Eggertsen G, Fey GH (1988). "Relationships between the gene and protein structure in human complement component C9". Biochemistry. 27 (17): 6529–6534. doi:10.1021/bi00417a050. PMID 3219351.
- ↑ Stanley KK, Luzio JP, Tschopp J, Kocher HP, Jackson P (1985). "The sequence and topology of human complement component C9". EMBO J. 4 (2): 375–382. PMC 554196
 . PMID 4018030.
. PMID 4018030. - ↑ Fey GH, Hugli TE, Podack ER, Gehring MR, Kan CC, DiScipio RG (1984). "Nucleotide sequence of cDNA and derived amino acid sequence of human complement component C9". Proc. Natl. Acad. Sci. U.S.A. 81 (23): 7298–7302. doi:10.1073/pnas.81.23.7298. PMC 392133
 . PMID 6095282.
. PMID 6095282. - ↑ Tschopp J, Masson D, Stanley KK (1986). "Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis". Nature. 322 (6082): 831–4. doi:10.1038/322831a0. PMID 2427956.
- ↑ Carlos J. Rosado; Ashley M. Buckle; Ruby H. P. Law; Rebecca E. Butcher; Wan-Ting Kan; Catherina H. Bird; Kheng Ung; Kylie A. Browne; Katherine Baran; Tanya A. Bashtannyk-Puhalovich; Noel G. Faux; Wilson Wong; Corrine J. Porter; Robert N. Pike; Andrew M. Ellisdon; Mary C. Pearce; Stephen P. Bottomley; Jonas Emsley; A. Ian Smith; Jamie Rossjohn; Elizabeth L. Hartland; Ilia Voskoboinik; Joseph A. Trapani; Phillip I. Bird; Michelle A. Dunstone & James C. Whisstock (2007). "A Common Fold Mediates Vertebrate Defense and Bacterial Attack". Science. 317 (5844): 1548–51. doi:10.1126/science.1144706. PMID 17717151.
- ↑ Hadders, MA (2012). "Assembly and regulation of the membrane attack complex based on structures of C5b6 and sC5b9". Cell Rep. 1. doi:10.1016/j.celrep.2012.02.003.
- ↑ Ronald Hoffman, Leslie E. Silberstein, Helen Heslop, Jeffrey Weitz, Hematology: Basic Principles and Practice, 6th ed., Elsevier, 2013, page 231.
- ↑ Abul K. Abbas, Andrew H. H. Lichtman, Shiv Pillai, Basic Immunology: Functions and Disorders of the Immune System, 5th ed., Elsevier, 2015, page 179.
External links
- Complement Membrane Attack Complex at the US National Library of Medicine Medical Subject Headings (MeSH)
- http://www.lib.mcg.edu/edu/esimmuno/ch2/lysis.htm