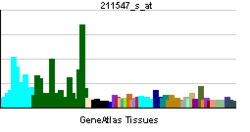
PAFAH1B1
| View/Edit Human | View/Edit Mouse |
Platelet-activating factor acetylhydrolase IB subunit alpha is an enzyme that in humans is encoded by the PAFAH1B1 gene.[3][4][5] The protein is often referred to as Lis1 and plays an important role in regulating the motor protein Dynein.[6]
Function
PAFAH1B1 was identified as encoding a gene that when mutated or lost caused the lissencephaly associated with Miller-Dieker syndrome. PAFAH1B1 encodes the non-catalytic alpha subunit of the intracellular Ib isoform of platelet-activating factor acteylhydrolase, a heterotrimeric enzyme that specifically catalyzes the removal of the acetyl group at the SN-2 position of platelet-activating factor (identified as 1-O-alkyl-2-acetyl-sn-glyceryl-3-phosphorylcholine). Two other isoforms of intracellular platelet-activating factor acetylhydrolase exist: one composed of multiple subunits, the other, a single subunit. In addition, a single-subunit isoform of this enzyme is found in serum.[5]
According to one study, PAFAH1B1 interacts with VLDLR receptor activated by reelin.[7]
Genomics
The gene is located at chromosome 17p13.3 on the Watson (plus) strand. The gene is 91,953 bases in length and encodes a protein of 410 amino acids (predicted molecular weight 46.638 kiloDaltons).
Interactions
PAFAH1B1 has been shown to interact with DYNC1H1,[8] CLIP1,[9] NDEL1,[10][11] NDE1,[12] PAFAH1B3,[13] PAFAH1B2,[13] NUDC,[14] TUBA1A[15] and Doublecortin.[16]
See also
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH (Aug 1993). "Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats". Nature. 364 (6439): 717–21. doi:10.1038/364717a0. PMID 8355785.
- ↑ Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH (Feb 1997). "Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome". Human Molecular Genetics. 6 (2): 157–64. doi:10.1093/hmg/6.2.157. PMID 9063735.
- 1 2 "Entrez Gene: PAFAH1B1 platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa".
- ↑ Kardon JR, Vale RD (Dec 2009). "Regulators of the cytoplasmic dynein motor". Nature Reviews Molecular Cell Biology. 10 (12): 854–65. doi:10.1038/nrm2804. PMID 19935668.
- ↑ Zhang G, Assadi AH, McNeil RS, Beffert U, Wynshaw-Boris A, Herz J, Clark GD, D'Arcangelo G (2007). Mueller U, ed. "The Pafah1b complex interacts with the reelin receptor VLDLR". PLOS ONE. 2 (2): e252. doi:10.1371/journal.pone.0000252. PMC 1800349
 . PMID 17330141.
. PMID 17330141. - ↑ Tai CY, Dujardin DL, Faulkner NE, Vallee RB (Mar 2002). "Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function". The Journal of Cell Biology. 156 (6): 959–68. doi:10.1083/jcb.200109046. PMC 2173479
 . PMID 11889140.
. PMID 11889140. - ↑ Coquelle FM, Caspi M, Cordelières FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, De Mey JR, Reiner O (May 2002). "LIS1, CLIP-170's key to the dynein/dynactin pathway". Molecular and Cellular Biology. 22 (9): 3089–102. doi:10.1128/MCB.22.9.3089-3102.2002. PMC 133759
 . PMID 11940666.
. PMID 11940666. - ↑ Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A (Jul 2003). "14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome". Nature Genetics. 34 (3): 274–85. doi:10.1038/ng1169. PMID 12796778.
- ↑ Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH (Dec 2000). "NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein". Neuron. 28 (3): 697–711. doi:10.1016/S0896-6273(00)00147-1. PMID 11163260.
- ↑ Efimov VP, Morris NR (Aug 2000). "The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein". The Journal of Cell Biology. 150 (3): 681–8. doi:10.1083/jcb.150.3.681. PMC 2175200
 . PMID 10931877.
. PMID 10931877. - 1 2 Sweeney KJ, Clark GD, Prokscha A, Dobyns WB, Eichele G (Apr 2000). "Lissencephaly associated mutations suggest a requirement for the PAFAH1B heterotrimeric complex in brain development". Mechanisms of Development. 92 (2): 263–71. doi:10.1016/S0925-4773(00)00242-2. PMID 10727864.
- ↑ Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee LY (May 1998). "The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC". Current Biology. 8 (10): 603–6. doi:10.1016/S0960-9822(98)70232-5. PMID 9601647.
- ↑ Sapir T, Elbaum M, Reiner O (Dec 1997). "Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit". The EMBO Journal. 16 (23): 6977–84. doi:10.1093/emboj/16.23.6977. PMC 1170301
 . PMID 9384577.
. PMID 9384577. - ↑ Caspi M, Atlas R, Kantor A, Sapir T, Reiner O (Sep 2000). "Interaction between LIS1 and doublecortin, two lissencephaly gene products". Human Molecular Genetics. 9 (15): 2205–13. doi:10.1093/oxfordjournals.hmg.a018911. PMID 11001923.
Further reading
- Tjoelker LW, Eberhardt C, Wilder C, Dietsch G, Trong HL, Cousens LS, Zimmerman GA, McIntyre TM, Stafforini DM, Prescott SM, Gray PW (1997). "Functional and structural features of plasma platelet-activating factor acetylhydrolase". Advances in Experimental Medicine and Biology. 416: 107–11. doi:10.1007/978-1-4899-0179-8_19. PMID 9131135.
- Stafforini DM, McIntyre TM, Zimmerman GA, Prescott SM (Jul 1997). "Platelet-activating factor acetylhydrolases". The Journal of Biological Chemistry. 272 (29): 17895–8. doi:10.1074/jbc.272.29.17895. PMID 9218411.
- Yamada Y, Yokota M (May 1998). "Roles of plasma platelet-activating factor acetylhydrolase in allergic, inflammatory, and atherosclerotic diseases". Japanese Circulation Journal. 62 (5): 328–35. doi:10.1253/jcj.62.328. PMID 9626899.
- Reiner O, Cahana A, Escamez T, Martinez S (2002). "LIS1-no more no less". Molecular Psychiatry. 7 (1): 12–6. doi:10.1038/sj/mp/4000975. PMID 11803439.
- Guerrini R, Carrozzo R (Apr 2002). "Epileptogenic brain malformations: clinical presentation, malformative patterns and indications for genetic testing". Seizure. 11 Suppl A: 532–43; quiz 544–7. doi:10.1053/seiz.2001.0650. PMID 12185771.
- Wynshaw-Boris A (Oct 2007). "Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development". Clinical Genetics. 72 (4): 296–304. doi:10.1111/j.1399-0004.2007.00888.x. PMID 17850624.
- Mizuguchi M, Takashima S, Kakita A, Yamada M, Ikeda K (Oct 1995). "Lissencephaly gene product. Localization in the central nervous system and loss of immunoreactivity in Miller-Dieker syndrome". The American Journal of Pathology. 147 (4): 1142–51. PMC 1870994
 . PMID 7573359.
. PMID 7573359. - Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K (Jul 1994). "Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected]". Nature. 370 (6486): 216–8. doi:10.1038/370216a0. PMID 8028668.
- Reiner O, Bar-Am I, Sapir T, Shmueli O, Carrozzo R, Lindsay EA, Baldini A, Ledbetter DH, Cahana A (Nov 1995). "LIS2, gene and pseudogene, homologous to LIS1 (lissencephaly 1), located on the short and long arms of chromosome 2". Genomics. 30 (2): 251–6. doi:10.1006/geno.1995.9880. PMID 8586424.
- Isumi H, Takashima S, Kakita A, Yamada M, Ikeda K, Mizuguchi M (Jan 1997). "Expression of the LIS-1 gene product in brain anomalies with a migration disorder". Pediatric Neurology. 16 (1): 42–4. doi:10.1016/S0887-8994(96)00260-3. PMID 9044400.
- Sapir T, Elbaum M, Reiner O (Dec 1997). "Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit". The EMBO Journal. 16 (23): 6977–84. doi:10.1093/emboj/16.23.6977. PMC 1170301
 . PMID 9384577.
. PMID 9384577. - Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee LY (May 1998). "The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC". Current Biology. 8 (10): 603–6. doi:10.1016/S0960-9822(98)70232-5. PMID 9601647.
- Pilz DT, Kuc J, Matsumoto N, Bodurtha J, Bernadi B, Tassinari CA, Dobyns WB, Ledbetter DH (Sep 1999). "Subcortical band heterotopia in rare affected males can be caused by missense mutations in DCX (XLIS) or LIS1". Human Molecular Genetics. 8 (9): 1757–60. doi:10.1093/hmg/8.9.1757. PMID 10441340.
- Sapir T, Cahana A, Seger R, Nekhai S, Reiner O (Oct 1999). "LIS1 is a microtubule-associated phosphoprotein". European Journal of Biochemistry / FEBS. 265 (1): 181–8. doi:10.1046/j.1432-1327.1999.00711.x. PMID 10491172.
- Sweeney KJ, Clark GD, Prokscha A, Dobyns WB, Eichele G (Apr 2000). "Lissencephaly associated mutations suggest a requirement for the PAFAH1B heterotrimeric complex in brain development". Mechanisms of Development. 92 (2): 263–71. doi:10.1016/S0925-4773(00)00242-2. PMID 10727864.