Salterprovirus
| Salterprovirus | |
|---|---|
 | |
| Halovirus His1 | |
| Virus classification | |
| Group: | Group I (dsDNA) |
| Genus: | Salterprovirus |
| Type Species | |
| |
Salterprovirus is a genus of viruses[1] that infect extremely halophilic archaea (isolated from haloarcula hispanica. There is currently only one species in this genus: the type species His 1 virus,[2][3] though His 2 virus (as yet unlisted by ICTV) is often discussed, even grouped, alongside His1V.[1][2] The genus name is derived from salt terminal protein virus, as their linear dsDNA genomes have proteins attached to the 5' termini (i.e. terminal proteins). This virus morphotype is commonly observed in hypersaline waters around the world, and salterproviruses may represent an environmentally common halovirus.
Taxonomy
The capsid is similar in shape to that of viruses infecting thermophilic Archaea, the Fuselloviridae, and His1V was originally described as a probable member of that group.[4] However, it was later found that there is no genetic relationship and their replication strategies are entirely different, and so they were reclassified into a new group, Salterprovirus.
Structure
Viruses in Salterprovirus are enveloped, with lemon-shaped geometries. Genomes are linear, around 145kb in length. The genome has 35 open reading frames.[2]
| Genus | Structure | Symmetry | Capsid | Genomic Arrangement | Genomic Segmentation |
|---|---|---|---|---|---|
| Salterprovirus | Lemon-shaped | Enveloped | Linear | Monopartite |
Life Cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by virus attaches to host cell. DNA-templated transcription is the method of transcription. Archaea and haloarcula hispanica serve as the natural host. Transmission routes are passive diffusion.[2]
| Genus | Host Details | Tissue Tropism | Entry Details | Release Details | Replication Site | Assembly Site | Transmission |
|---|---|---|---|---|---|---|---|
| Salterprovirus | Archea: Haloarcula hispanica | None | Injection | Unknown | Cytoplasm | Cytoplasm | Passive diffusion |
Discussion of His1V and His2V
The genomes of His1 and His2 are similar in size (14,464 bp & 16,067 bp, respectively) and have the same number of predicted open reading frames (35), but they share almost no nucleotide sequence similarity. However, at the protein sequence level, their DNA polymerases share 42% identity. This, together with their similar morphologies, DNA structure (length, inverted terminal repeat sequences, terminal proteins), and mode of replication indicate that they ought to belong to the same virus group.
A picture of His1 particles can be viewed at the www.haloarchaea.com website here, and scroll down. Curiously, homologues of the genes for the capsid proteins of His2 are widely distributed in genomes of haloarchaea.[5] The genomic loci containing these also tend to have other virus/plasmid genes nearby, and the term ViPREs[6] (for Virus and Plasmid Related Elements) has been proposed to denote these gene clusters, which may parallel the situation in thermophilic Archaea, where plasmids and viruses are closely linked.
Transfection of host cells (Haloarcula hispanica) by His2 DNA was demonstrated by Porter et al. (2008),[7] who showed that infectious virus could be produced from viral DNA as long as the terminal proteins were not damaged by protease treatment. This is consistent with their likely role in DNA replication. It could then be shown that His2 DNA could transfect a wide variety of haloarchaeal species even though the cells were not susceptible to infection by His2 virus. In these experiments, transfected cells were plated along with cells of the host, Haloarcula hispanica, and virus produced by the transfectants was detected by infection of nearby cells of the susceptible host species. The experiments showed that the cytoplasms of many haloarchaea are able to replicate virus as long as the DNA (with terminal proteins) can enter. It is the virus capsid that determines cell selection, most likely by proteins in the short tail at one end of the spindle-shaped virion.
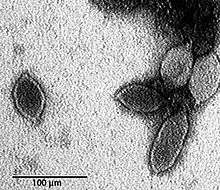
Once transfection had been established, it was possible to develop a system for mutagenesis, and the same publication described the use of an in vitro transposon system for inserting DNA randomly into the His2 genome. Transfection of the reaction products recovered virus recombinants with insertions, and mapping of these located regions of the His2 genome that were non-essential. Most insertions were found to lie in one of the two inverted terminal repeat (ITR) sequences. A few occurred at the very ends of three open reading frames. The study proved you could create His2 mutants, that you could insert at least 435 extra bp into the genome, and you could recover viable mutants with these insertions. This opens the way to analysing the functions of each of the virus genes, what they do and how they interact with each other and with the host cell. At present, almost all of the predicted virus proteins have no known function. It is not clear how much extra DNA can be inserted into the genome, but as is evident from the electronmicrograph, capsid size may not be a limiting factor, as the particle sizes of His1 vary considerably in length (and so probably for His2, although negative-stain EM of these particles is much more difficult).
References
- 1 2 Bath C, Cukalac T, Porter K, Dyall-Smith ML. "His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus genus, Salterprovirus" Virology. 2006 350:228-39
- 1 2 3 4 "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ↑ ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ↑ Bath C, Dyall-Smith, ML. "His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica" J. Virol. 1998 72:9392-9395
- ↑ Bath C, Cukalac T, Porter K, Dyall-Smith ML. "His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus" Virology. 2006 350:228-39
- ↑ Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, et al. (2011) Haloquadratum walsbyi : Limited Diversity in a Global Pond. PLoS ONE. 6(6): e20968. doi:10.1371/journal.pone.0020968
- ↑ Porter K, Dyall-Smith ML. "Transfection of haloarchaea by the DNAs of spindle and round haloviruses and the use of transposon mutagenesis to identify non-essential regions." Molecular Microbiology. 2008. 70:1236-45