Cancer stem cell
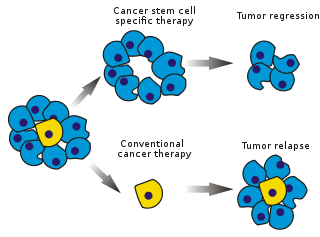
Cancer stem cells (CSCs) are cancer cells (found within tumors or hematological cancers) that possess characteristics associated with normal stem cells, specifically the ability to give rise to all cell types found in a particular cancer sample. CSCs are therefore tumorigenic (tumor-forming), perhaps in contrast to other non-tumorigenic cancer cells. CSCs may generate tumors through the stem cell processes of self-renewal and differentiation into multiple cell types. Such cells are hypothesized to persist in tumors as a distinct population and cause relapse and metastasis by giving rise to new tumors. Therefore, development of specific therapies targeted at CSCs holds hope for improvement of survival and quality of life of cancer patients, especially for patients with metastatic disease.
Existing cancer treatments have mostly been developed based on animal models, where therapies able to promote tumor shrinkage were deemed effective. However, animals do not provide a complete model of human disease. In particular, in mice, whose life spans do not exceed two years, tumor relapse is difficult to study.
The efficacy of cancer treatments is, in the initial stages of testing, often measured by the ablation fraction of tumor mass (fractional kill). As CSCs form a small proportion of the tumor, this may not necessarily select for drugs that act specifically on the stem cells. The theory suggests that conventional chemotherapies kill differentiated or differentiating cells, which form the bulk of the tumor but do not generate new cells. A population of CSCs, which gave rise to it, could remain untouched and cause relapse.
Cancer stem cells were first identified by John Dick in acute myeloid leukemia in the late 1990s. Since the early 2000s they have been an intense cancer research focus.[1]
Tumor propagation models
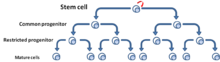
In different tumor subtypes, cells within the tumor population exhibit functional heterogeneity and tumors are formed from cells with various proliferative and differentiation capacities.[2] This functional heterogeneity among cancer cells has led to the creation of multiple propagation models to account for heterogeneity and differences in tumor-regenerative capacity: the cancer stem cell (CSC) and clonal evolution models[3]
The cancer stem cell model refers to a subset of tumor cells that have pluripotency and are able to generate the diverse tumor cells.[3] These cells have been termed cancer stem cells to reflect their stem-like properties. One implication of the CSC model and the existence of CSCs is that the tumor population is hierarchically arranged with CSCs lying at the apex[4] (Fig. 3).
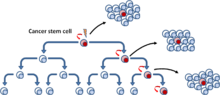
The clonal evolution model postulates that mutant tumor cells with a growth advantage outproliferate others. Cells in the dominant population have a similar potential for initiating tumor growth[5] (Fig. 4).

[6] These two models are not mutually exclusive, as CSCs themselves undergo clonal evolution. Thus, the secondary more dominant CSCs may emerge, if a mutation confers more aggressive properties[7] (Fig. 5).

Debate
The existence of CSCs is under debate, because many studies found no cells with their specific characteristics.[8] Cancer cells must be capable of continuous proliferation and self-renewal to retain the many mutations required for carcinogenesis and to sustain the growth of a tumor, since differentiated cells (constrained by the Hayflick Limit[9]) cannot divide indefinitely. If most tumor cells are endowed with stem cell properties, targeting tumor size directly is a valid strategy. If they are a small minority, targeting them may be more effective. Another debate is over the origin of CSCs - whether from disregulation of normal stem cells or from a more specialized population that acquired the ability to self-renew (which is related to the issue of stem cell plasticity).
Evidence
The first conclusive evidence for CSCs came in 1997. Bonnet and Dick isolated a subpopulation of leukemia cells that expressed surface marker CD34, but not CD38.[10] The authors established that the CD34+/CD38− subpopulation is capable of initiating tumors in NOD/SCID mice that were histologically similar to the donor. The first evidence of a solid tumor cancer stem-like cell followed in 2002 with the discovery of a clonogenic, sphere-forming cell isolated and characterized from human brain gliomas. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro.[11]
In cancer research experiments, tumor cells are sometimes injected into an experimental animal to establish a tumor. Disease progression is then followed in time and novel drugs can be tested for their efficacy. Tumor formation requires thousands or tens of thousands of cells to be introduced. Classically, this was explained by poor methodology (i.e., the tumor cells lose their viability during transfer) or the critical importance of the microenvironment, the particular biochemical surroundings of the injected cells. Supporters of the CSC paradigm argue that only a small fraction of the injected cells, the CSCs, have the potential to generate a tumor. In human acute myeloid leukemia the frequency of these cells is less than 1 in 10,000.[10]
Further evidence comes from histology. Many tumors are heterogeneous and contain multiple cell types native to the host organ. Heterogeneity is commonly retained by tumor metastases. This suggests that the cell that produced them had the capacity to generate multiple cell types, a classical hallmark of stem cells.[10]
The existence of leukemia stem cells prompted research into other cancers. CSCs have recently been identified in several solid tumors, including:
- Brain[12]
- Breast[13]
- Colon[14]
- Ovary[15][16][17]
- Pancreas[18]
- Prostate[19][20]
- Melanoma[21][22][23][24]
- Multiple Myeloma[25][26]
- Non-melanoma skin cancer[27][28]
Mechanistic and mathematical models
Once the pathways to cancer are hypothesized, it is possible to develop predictive mathematical models,[29] e.g., based on the cell compartment method. For instance, the growths of abnormal cells can be denoted with specific mutation probabilities. Such a model predicted that repeated insult to mature cells increases the formation of abnormal progeny and the risk of cancer.[30] The clinical efficacy of such models[31] remains unestablished.
Origin

The origin of CSCs is an active research area. The answer may depend on the tumor type and phenotype. So far the hypothesis that tumors originate from a single "cell of origin" has not been demonstrated using the cancer stem cell model. This is because cancer stem cells are not present in end-stage tumors.
Origin hypotheses include mutants in developing stem or progenitor cells, mutants in adult stem cells or adult progenitor cells and mutant, differentiated cells that acquire stem-like attributes. These theories often focus on a tumor's "cell of origin".
Hypotheses
Stem cell mutation
The "mutation in stem cell niche populations during development" hypothesis claims that these developing stem populations are mutated and then reproduce so that the mutation is shared by many descendants. These daughter cells are much closer to becoming tumors and their numbers increase the chance of a cancerous mutation.[32]
Adult stem cells
Another theory associates adult stem cells (ASC) with tumor formation. This is most often associated with tissues with a high rate of cell turnover (such as the skin or gut). In these tissues, ASCs are candidates because of their frequent cell divisions (compared to most ASCs) in conjunction with the long lifespan of ASCs. This combination creates the ideal set of circumstances for mutations to accumulate: mutation accumulation is the primary factor that drives cancer initiation. Evidence shows that the association represents an actual phenomenon, although specific cancers have been linked to a specific cause.[33][34]
De-differentiation
De-differentiation of mutated cells may create stem cell-like characteristics, suggesting that any cell might become a cancer stem cell. In other words, a fully differentiated cell undergoes several mutations that drive it back to a stem-like state.
Hierarchy
The concept of tumor hierarchy claims that a tumor is a heterogeneous population of mutant cells, all of which share some mutations, but vary in specific phenotype. A tumor hosts several types of stem cells, one optimal to the specific environment and other less successful lines. These secondary lines may be more successful in other environments, allowing the tumor to adapt, including adaptation to therapeutic intervention. If correct, this concept impacts cancer stem cell-specific treatment regimes.[35] Such a hierarchy would complicate attempts to pinpoint the origin.
Identification
CSCs, now reported in most human tumors, are commonly identified and enriched using strategies for identifying normal stem cells that are similar across studies.[36] These procedures include fluorescence-activated cell sorting (FACS), with antibodies directed at cell-surface markers and functional approaches including side population assay or Aldefluor assay.[37] The CSC-enriched result is then implanted, at various doses, in immune-deficient mice to assess its tumor development capacity. This in vivo assay is called a limiting dilution assay. The tumor cell subsets that can initiate tumor development at low cell numbers are further tested for self-renewal capacity in serial tumor studies.[38]
CSC can also be identified by efflux of incorporated Hoechst dyes via multidrug resistance (MDR) and ATP-binding cassette (ABC) Transporters.[37]
Another approach is sphere-forming assays. Many normal stem cells such as hematopoietics or stem cells from tissues, under special culture conditions, form three-dimensional spheres that can differentiate. As with normal stem cells, the CSCs isolated from brain or prostate tumors also have the ability to form anchor-independent spheres.[39]
Heterogeneity (markers)
CSCs have been identified in various solid tumors. Markers specific for normal stem cells are commonly used for isolating CSCs from solid and hematological tumors. Cell surface markers have proved useful for isolation of CSC-enriched populations including CD133 (also known as PROM1), CD44, CD24, EpCAM (epithelial cell adhesion molecule, also known as epithelial specific antigen, ESA), THY1, ATP-binding cassette B5 (ABCB5).,[40] and CD200.
CD133 (prominin 1) is a five-transmembrane domain glycoprotein expressed on CD34+ stem and progenitor cells, in endothelial precursors and fetal neural stem cells. It has been detected using its glycosylated epitope known as AC133.
EpCAM (epithelial cell adhesion molecule, ESA, TROP1) is hemophilic Ca2+-independent cell adhesion molecule expressed on the basolateral surface of most epithelial cells.
CD90 (THY1) is a glycosylphosphatidylinositol glycoprotein anchored in the plasma membrane and involved in signal transduction. It may also mediate adhesion between thymocytes and thymic stroma.
CD44 (PGP1) is an adhesion molecule that has pleiotropic roles in cell signaling, migration and homing. It has multiple isoforms, including CD44H, which exhibits high affinity for hyaluronate and CD44V which has metastatic properties.
CD24 (HSA) is a glycosylated glycosylphosphatidylinositol-anchored adhesion molecule, which has co-stimulatory role in B and T cells.
CD200 (OX-2) is a type 1 membrane glycoprotein, which delivers an inhibitory signal to immune cells including T cells, natural killer cells and macrophages.
ALDH is a ubiquitous aldehyde dehydrogenase family of enzymes, which catalyzes the oxidation of aromatic aldehydes to carboxyl acids. For instance, it has a role in conversion of retinol to retinoic acid, which is essential for survival.[41][42]
The first solid malignancy from which CSCs were isolated and identified was breast cancer and they are the most intensely studied. Breast CSCs have been enriched in CD44+CD24−/low,[40] SP[43] and ALDH+ subpopulations.[44][45] Breast CSCs are apparently phenotypically diverse. CSC marker expression in breast cancer cells is apparently heterogeneous and breast CSC populations vary across tumors.[46] Both CD44+CD24− and CD44+CD24+ cell populations are tumor initiating cells; however, CSC are most highly enriched using the marker profile CD44+CD49fhiCD133/2hi.[47]
CSCs have been reported in many brain tumors. Stem-like tumor cells have been identified using cell surface markers including CD133,[48] SSEA-1 (stage-specific embryonic antigen-1),[49] EGFR[50] and CD44.[51] The use of CD133 for identification of brain tumor stem-like cells may be problematic because tumorigenic cells are found in both CD133+ and CD133− cells in some gliomas and some CD133+ brain tumor cells may not possess tumor-initiating capacity.[50]
CSCs were reported in human colon cancer.[52] For their identification, cell surface markers such as CD133,[52] CD44[53] and ABCB5,[54] functional analysis including clonal analysis [55] and Aldefluor assay were used.[56] Using CD133 as a positive marker for colon CSCs generated conflicting results. The AC133 epitope, but not the CD133 protein, is specifically expressed in colon CSCs and its expression is lost upon differentiation.[57] In addition, CD44+ colon cancer cells and additional sub-fractionation of CD44+EpCAM+ cell population with CD166 enhance the success of tumor engraftments.[53]
Multiple CSCs have been reported in prostate,[58] lung and many other organs, including liver, pancreas, kidney or ovary.[41][59] In prostate cancer, the tumor-initiating cells have been identified in CD44+[60] cell subset as CD44+α2β1+,[61] TRA-1-60+CD151+CD166+ [62] or ALDH+ [63] cell populations. Putative markers for lung CSCs have been reported, including CD133+,[64] ALDH+,[65] CD44+ [66] and oncofetal protein 5T4+.[67]
Metastasis
Metastasis is the major cause of tumor lethality. However, not every tumor cell can metastasize. This potential depends on factors that determine growth, angiogenesis, invasion and other basic processes.
Epithelial-mesenchymal transition
In epithelial tumors, the epithelial-mesenchymal transition (EMT) is considered to be a crucial event.[68] EMT and the reverse transition from mesenchymal to an epithelial phenotype (MET) are involved in embryonic development, which involves disruption of epithelial cell homeostasis and the acquisition of a migratory mesenchymal phenotype.[69] EMT appears to be controlled by canonical pathways such as WNT and transforming growth factor β.[70]
EMT's important feature is the loss of membrane E-cadherin in adherens junctions, where β-catenin may play a significant role. Translocation of β-catenin from adherens junctions to the nucleus may lead to a loss of E-cadherin and subsequently to EMT. Nuclear β-catenin apparently can directly, transcriptionally activate EMT-associated target genes, such as the E-cadherin gene repressor SLUG (also known as SNAI2).[71] Mechanical properties of the tumor microenvironment, or hypoxia, can influence CSC properties and metastatic behavior.[72]
Tumor cells undergoing an EMT may be precursors for metastatic cancer cells, or even metastatic CSCs.[73] In the invasive edge of pancreatic carcinoma, a subset of CD133+CXCR4+ (receptor for CXCL12 chemokine also known as a SDF1 ligand) cells was defined. These cells exhibited significantly stronger migratory activity than their counterpart CD133+CXCR4− cells, but both showed similar tumor development capacity.[74] Moreover, inhibition of the CXCR4 receptor reduced metastatic potential without altering tumorigenic capacity.[75]
Two-phase expression pattern
In breast cancer CD44+CD24−/low cells are detectable in metastatic pleural effusions.[40] By contrast, an increased number of CD24+ cells have been identified in distant metastases in breast cancer patients.[76] It is possible that CD44+CD24−/low cells initially metastasize and in the new site change their phenotype and undergo limited differentiation.[77] The two-phase expression pattern hypothesis proposes two forms of cancer stem cells - stationary (SCS) and mobile (MCS). SCS are embedded in tissue and persist in differentiated areas throughout tumor progression. MCS are located at the tumor-host interface. These cells are apparently derived from SCS through the acquisition of transient EMT (Figure 7).[78]

Implications
CSCs have implications for cancer therapy, including for disease identification, selective drug targets, prevention of metastasis and intervention strategies.
Treatment
Somatic stem cells are naturally resistant to chemotherapeutic agents. They produce various pumps (such as MDR) that pump out drugs and DNA repair proteins. They have a slow rate of cell turnover (chemotherapeutic agents naturally target rapidly replicating cells). CSCs that develop from normal stem cells may also produce these proteins, which could increase their resistance towards chemotherapy. The surviving CSCs then repopulate the tumor, causing a relapse.[79]
Targeting
Selectively targeting CSCs may allow treatment of aggressive, non-resectable tumors, as well as prevent metastasis and relapse.[79] The hypothesis suggests that upon CSC elimination, cancer could regress due to differentiation and/or cell death. The fraction of tumor cells that are CSCs and therefore need to be eliminated is unclear.[80]
Studies looked for specific markers[13] and for proteomic and genomic tumor signatures that distinguish CSCs from others.[81] In 2009, scientists identified the compound salinomycin, which selectively reduces the proportion of breast CSCs in mice by more than 100-fold relative to Paclitaxel, a commonly used chemotherapeutic agent.[82] Some types of cancer cells can survive treatment with salinomycin through autophagy,[83] whereby cells use acidic organelles such as lysosomes to degrade and recycle certain types of proteins. The use of autophagy inhibitors can kill cancer stem cells that survive by autophagy.[84]
The cell surface receptor interleukin-3 receptor-alpha (CD123) is overexpressed on CD34+CD38- leukemic stem cells (LSCs) in acute myelogenous leukemia (AML) but not on normal CD34+CD38- bone marrow cells.[85] Treating AML-engrafted NOD/SCID mice with a CD123-specific monoclonal antibody impaired LSCs homing to the bone marrow and reduced overall AML cell repopulation including the proportion of LSCs in secondary mouse recipients.[86]
A 2015 study packaged nanoparticles with miR-34a and ammonium bicarbonate and delivered them to prostate CSCs in a mouse model. Then they irradiated the area with near-infrared laser light. This caused the nanoparticles to swell three times or more in size bursting the endosomes and dispersing the RNA in the cell. miR-34a can lower the levels of CD44.[87][88]
Pathways
The design of new drugs for targeting CSCs requires understanding the cellular mechanisms that regulate cell proliferation. The first advances in this area were made with hematopoietic stem cells (HSCs) and their transformed counterparts in leukemia, the disease for which the origin of CSCs is best understood. Stem cells of many organs share the same cellular pathways as leukemia-derived HSCs.
A normal stem cell may be transformed into a CSC through disregulation of the proliferation and differentiation pathways controlling it or by inducing oncoprotein activity.
BMI-1
The Polycomb group transcriptional repressor Bmi-1 was discovered as a common oncogene activated in lymphoma[89] and later shown to regulate HSCs.[90] The role of Bmi-1 has been illustrated in neural stem cells.[91] The pathway appears to be active in CSCs of pediatric brain tumors.[92]
Notch
The Notch pathway plays a role in controlling stem cell proliferation for several cell types including hematopoietic, neural and mammary[93] SCs. Components of this pathway have been proposed to act as oncogenes in mammary[94] and other tumors.
A branch of the Notch signaling pathway that involves the transcription factor Hes3 regulates a number of cultured cells with CSC characteristics obtained from glioblastoma patients.[95]
Sonic hedgehog and Wnt
These developmental pathways are SC regulators.[96] Both Sonic hedgehog (SHH) and Wnt pathways are commonly hyperactivated in tumors and are necessary to sustain tumor growth. However, the Gli transcription factors that are regulated by SHH take their name from gliomas, where they are highly expressed. A degree of crosstalk exists between the two pathways and they are commonly activated together.[97] By contrast, in colon cancer hedgehog signalling appears to antagonise Wnt.[98]
Sonic hedgehog blockers are available, such as cyclopamine. A water-soluble cyclopamine may be more effective in cancer treatment. DMAPT, a water-soluble derivative of parthenolide, induces oxidative stress and inhibits NF-κB signaling[99] for AML (leukemia) and possibly myeloma and prostate cancer. Telomerase is a study subject in CSC physiology.[100] GRN163L (Imetelstat) was recently started in trials to target myeloma stem cells.
Wnt signaling can become independent of regular stimuli, through mutations in downstream oncogenes and tumor suppressor genes that become permanently activated even though the normal receptor has not received a signal. β-catenin binds to transcription factors such as the protein TCF4 and in combination the molecules activate the necessary genes. LF3 strongly inhibits this binding in vitro, in cell lines and reduced tumor growth in mouse models. It prevented replication and reduced their ability to migrate, all without affecting healthy cells. No cancer stem cells remained after treatment. The discovery was the product of "rational drug design", involving AlphaScreens and ELISA technologies.[101]
References
- ↑ Mukherjee, Siddhartha. "The Cancer Sleeper Cell". New York Times. New York Times. Retrieved 15 July 2014.
- ↑ Heppner, GH; Miller, BE (1983). "Tumor heterogeneity: biological implications and therapeutic consequences". Cancer metastasis reviews. 2 (1): 5–23. doi:10.1007/BF00046903. PMID 6616442.
- 1 2 Reya, T; Morrison, SJ; Clarke, MF; Weissman, IL (Nov 1, 2001). "Stem cells, cancer, and cancer stem cells". Nature. 414 (6859): 105–11. doi:10.1038/35102167. PMID 11689955.
- ↑ Bonnet, D; Dick, JE (July 1997). "Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell". Nature Medicine. 3 (7): 730–7. doi:10.1038/nm0797-730. PMID 9212098.
- ↑ Barabé, F; Kennedy, JA; Hope, KJ; Dick, JE (Apr 27, 2007). "Modeling the initiation and progression of human acute leukemia in mice". Science. 316 (5824): 600–4. doi:10.1126/science.1139851. PMID 17463288.
- ↑ Nowell, PC (Oct 1, 1976). "The clonal evolution of tumor cell populations". Science. 194 (4260): 23–8. doi:10.1126/science.959840. PMID 959840.
- ↑ Clark, EA; Golub, TR; Lander, ES; Hynes, RO (Aug 3, 2000). "Genomic analysis of metastasis reveals an essential role for RhoC". Nature. 406 (6795): 532–5. doi:10.1038/35020106. PMID 10952316.
- ↑ Gupta PB, Chaffer CL, Weinberg RA (2009). "Cancer stem cells: mirage or reality?". Nat Med. 15 (9): 1010–2. doi:10.1038/nm0909-1010. PMID 19734877. >
- ↑ Hayflick L (1965). "The Limited in Vitro Lifetime of Human Diploid Cell Strains". Exp Cell Res. 37: 614–636. doi:10.1016/0014-4827(65)90211-9. PMID 14315085.
- 1 2 3 Bonnet D, Dick JE (July 1997). "Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell". Nature Medicine. 3 (7): 730–7. doi:10.1038/nm0797-730. PMID 9212098.
- ↑ Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA (Sep 2002). "Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro.". Glia. 39 (3): 193–206. doi:10.1002/glia.10094. PMID 12203386.
- ↑ Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB (September 2003). "Identification of a cancer stem cell in human brain tumors". Cancer Research. 63 (18): 5821–8. PMID 14522905.
- 1 2 Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (April 2003). "Prospective identification of tumorigenic breast cancer cells". Proceedings of the National Academy of Sciences of the United States of America. 100 (7): 3983–8. doi:10.1073/pnas.0530291100. PMC 153034
 . PMID 12629218.
. PMID 12629218. - ↑ O'Brien CA, Pollett A, Gallinger S, Dick JE (January 2007). "A human colon cancer cell capable of initiating tumour growth in immunodeficient mice". Nature. 445 (7123): 106–10. doi:10.1038/nature05372. PMID 17122772.
- ↑ Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP (June 2008). "Identification and characterization of ovarian cancer-initiating cells from primary human tumors". Cancer Research. 68 (11): 4311–20. doi:10.1158/0008-5472.CAN-08-0364. PMC 2553722
 . PMID 18519691.
. PMID 18519691. - ↑ "Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance.". Cell Cycle. 8 (1): 158–66. Jan 2009. doi:10.4161/cc.8.1.7533. PMID 19158483.
- ↑ Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, Silasi DA, Steffensen KD, Waldstrom M (Jan 2009). "Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance". Cell Cycle. 8 (1): 158–66. doi:10.4161/cc.8.1.7533. PMID 19158483.
- ↑ Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM (February 2007). "Identification of pancreatic cancer stem cells". Cancer Research. 67 (3): 1030–7. doi:10.1158/0008-5472.CAN-06-2030. PMID 17283135.
- ↑ Maitland NJ, Collins AT (June 2008). "Prostate cancer stem cells: a new target for therapy". J. Clin. Oncol. 26 (17): 2862–70. doi:10.1200/JCO.2007.15.1472. PMID 18539965.
- ↑ Lang Sh; Frame F; Collins A (January 2009). "Prostate cancer stem cells". J. Pathol. 217 (2): 299–306. doi:10.1002/path.2478. PMC 2673349
 . PMID 19040209.
. PMID 19040209. - ↑ Schatton T; Murphy GF; Frank, NY; Yamaura K; Waaga-Gasser AM; Gasser M; Zhan Q; Jordan S; Duncan LM; Weishaupt C; Fuhlbrigge RC; Kupper TS; Sayegh MH; Frank MH (Jan 2008). "Identification of cells initiating human melanomas". Nature. 451 (7176): 345–9. doi:10.1038/nature06489. PMC 3660705
 . PMID 18202660.
. PMID 18202660. - ↑ Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, Longaker MT, Weissman IL (Jul 2010). "Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271". Nature. 466 (7302): 133–7. doi:10.1038/nature09161. PMC 2898751
 . PMID 20596026.
. PMID 20596026. - ↑ Schmidt P, Kopecky C, Hombach A, Zigrino P, Mauch C, Abken H (Feb 2011). "Eradication of melanomas by targeted elimination of a minor subset of tumor cells". PNAS. 108 (6): 2474–9. doi:10.1073/pnas.1009069108. PMC 3038763
 . PMID 21282657.
. PMID 21282657. - ↑ Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, Seifert B, Moch H, Dummer R, van den Broek M, Sommer L (Mar 2011). "Human CD271-Positive Melanoma Stem Cells Associated with Metastasis Establish Tumor Heterogeneity and Long-Term Growth". Cancer Res. 71 (8): 3098–109. doi:10.1158/0008-5472.CAN-10-3997. PMID 21393506.
- ↑ Matsui W, Huff CA, Wang Q, et al. (March 2004). "Characterization of clonogenic multiple myeloma cells". Blood. 103 (6): 2332–6. doi:10.1182/blood-2003-09-3064. PMC 3311914
 . PMID 14630803.
. PMID 14630803. - ↑ Matsui W, Wang Q, Barber JP, et al. (January 2008). "Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance". Cancer Res. 68 (1): 190–7. doi:10.1158/0008-5472.CAN-07-3096. PMC 2603142
 . PMID 18172311.
. PMID 18172311. - ↑ Colmont CS, Benketah A, Reed SH, Hawk NV, Telford WG, Ohyama M, Udey MC, Yee CL, Vogel JC, Patel GK (Jan 2013). "CD200-expressing human basal cell carcinoma cells initiate tumor growth" (PDF). PNAS. 110 (4): 1434–9. doi:10.1073/pnas.1211655110. PMC 3557049
 . PMID 23292936.
. PMID 23292936. - ↑ Patel GK, Yee CL, Terunuma A, Telford WG, Voong N, Yuspa SH, Vogel JC (Feb 2012). "Identification and characterization of tumor-initiating cells in human primary cutaneous squamous cell carcinoma" (PDF). J Invest Dermatol. 132 (2): 401–409. doi:10.1038/jid.2011.317. PMC 3258300
 . PMID 22011906.
. PMID 22011906. - ↑ Preziosi, Luigi (2003). Cancer Modelling and Simulation. Boca Raton: CRC Press. ISBN 1-58488-361-8.
- ↑ Ganguly R, Puri IK (February 2006). "Mathematical model for the cancer stem cell hypothesis". Cell proliferation. 39 (1): 3–14. doi:10.1111/j.1365-2184.2006.00369.x. PMID 16426418.
- ↑ Ganguly R, Puri IK (June 2007). "Mathematical model for chemotherapeutic drug efficacy in arresting tumour growth based on the cancer stem cell hypothesis". Cell proliferation. 40 (3): 338–354. doi:10.1111/j.1365-2184.2007.00434.x. PMID 17531079.
- ↑ Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y (June 2009). "Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model". Cancer Cell. 15 (6): 514–26. doi:10.1016/j.ccr.2009.04.001. PMC 2721466
 . PMID 19477430.
. PMID 19477430. - ↑ López-Lázaro, Miguel (2015-01-01). "The migration ability of stem cells can explain the existence of cancer of unknown primary site. Rethinking metastasis". Oncoscience. 2 (5): 467–475. doi:10.18632/oncoscience.159. ISSN 2331-4737. PMC 4468332
 . PMID 26097879.
. PMID 26097879. - ↑ López-Lázaro, Miguel (2015-08-18). "Stem cell division theory of cancer". Cell Cycle (Georgetown, Tex.). 14 (16): 2547–2548. doi:10.1080/15384101.2015.1062330. ISSN 1551-4005. PMID 26090957.
- ↑ Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM (October 2006). "Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells". Cancer Research. 66 (19): 9339–44. doi:10.1158/0008-5472.CAN-06-3126. PMID 16990346.
- ↑ Golebiewska, A; Brons, NH; Bjerkvig, R; Niclou, SP (Feb 4, 2011). "Critical appraisal of the side population assay in stem cell and cancer stem cell research". Cell stem cell. 8 (2): 136–47. doi:10.1016/j.stem.2011.01.007. PMID 21295271.
- 1 2 Scharenberg, CW; Harkey, MA; Torok-Storb, B (Jan 15, 2002). "The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors". Blood. 99 (2): 507–12. doi:10.1182/blood.V99.2.507. PMID 11781231.
- ↑ Pastrana, E; Silva-Vargas, V; Doetsch, F (May 6, 2011). "Eyes wide open: a critical review of sphere-formation as an assay for stem cells". Cell stem cell. 8 (5): 486–98. doi:10.1016/j.stem.2011.04.007. PMC 3633588
 . PMID 21549325.
. PMID 21549325. - ↑ Nicolis, SK (February 2007). "Cancer stem cells and "stemness" genes in neuro-oncology". Neurobiology of disease. 25 (2): 217–29. doi:10.1016/j.nbd.2006.08.022. PMID 17141509.
- 1 2 3 Al-Hajj, M; Wicha, MS; Benito-Hernandez, A; Morrison, SJ; Clarke, MF (Apr 1, 2003). "Prospective identification of tumorigenic breast cancer cells". Proceedings of the National Academy of Sciences of the United States of America. 100 (7): 3983–8. doi:10.1073/pnas.0530291100. PMC 153034
 . PMID 12629218.
. PMID 12629218. - 1 2 Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA, Palle K, Rocconi RP (Sep 12, 2014). "ALDH1A1 Maintains Ovarian Cancer Stem Cell-Like Properties by Altered Regulation of Cell Cycle Checkpoint and DNA Repair Network Signaling.". PLOS ONE. 9 (9): e107142. doi:10.1371/journal.pone.0107142. PMC 4162571
 . PMID 25216266.
. PMID 25216266. - ↑ Visvader, JE; Lindeman, GJ (October 2008). "Cancer stem cells in solid tumours: accumulating evidence and unresolved questions". Nature Reviews Cancer. 8 (10): 755–68. doi:10.1038/nrc2499. PMID 18784658.
- ↑ Hirschmann-Jax, C; Foster, AE; Wulf, GG; Nuchtern, JG; Jax, TW; Gobel, U; Goodell, MA; Brenner, MK (Sep 28, 2004). "A distinct "side population" of cells with high drug efflux capacity in human tumor cells". Proceedings of the National Academy of Sciences of the United States of America. 101 (39): 14228–33. doi:10.1073/pnas.0400067101. PMC 521140
 . PMID 15381773.
. PMID 15381773. - ↑ Ginestier, C; Hur, MH; Charafe-Jauffret, E; Monville, F; Dutcher, J; Brown, M; Jacquemier, J; Viens, P; Kleer, CG; Liu, S; Schott, A; Hayes, D; Birnbaum, D; Wicha, MS; Dontu, G (November 2007). "ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome". Cell stem cell. 1 (5): 555–67. doi:10.1016/j.stem.2007.08.014. PMC 2423808
 . PMID 18371393.
. PMID 18371393. - ↑ Pece, S; Tosoni, D; Confalonieri, S; Mazzarol, G; Vecchi, M; Ronzoni, S; Bernard, L; Viale, G; Pelicci, PG; Di Fiore, PP (Jan 8, 2010). "Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content". Cell. 140 (1): 62–73. doi:10.1016/j.cell.2009.12.007. PMID 20074520.
- ↑ Deng, S; Yang, X; Lassus, H; Liang, S; Kaur, S; Ye, Q; Li, C; Wang, LP; Roby, KF; Orsulic, S; Connolly, DC; Zhang, Y; Montone, K; Bützow, R; Coukos, G; Zhang, L (Apr 21, 2010). Cao, Yihai, ed. "Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers". PLOS ONE. 5 (4): e10277. doi:10.1371/journal.pone.0010277. PMC 2858084
 . PMID 20422001.
. PMID 20422001. - ↑ Meyer, MJ; Fleming, JM; Lin, AF; Hussnain, SA; Ginsburg, E; Vonderhaar, BK (Jun 1, 2010). "CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer". Cancer Research. 70 (11): 4624–33. doi:10.1158/0008-5472.CAN-09-3619. PMID 20484027.
- ↑ Singh, SK; Hawkins, C; Clarke, ID; Squire, JA; Bayani, J; Hide, T; Henkelman, RM; Cusimano, MD; Dirks, PB (Nov 18, 2004). "Identification of human brain tumour initiating cells". Nature. 432 (7015): 396–401. doi:10.1038/nature03128. PMID 15549107.
- ↑ Son, MJ; Woolard, K; Nam, DH; Lee, J; Fine, HA (May 8, 2009). "SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma". Cell stem cell. 4 (5): 440–52. doi:10.1016/j.stem.2009.03.003. PMID 19427293.
- 1 2 Mazzoleni, S; Politi, LS; Pala, M; Cominelli, M; Franzin, A; Sergi Sergi, L; Falini, A; De Palma, M; Bulfone, A; Poliani, PL; Galli, R (Oct 1, 2010). "Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis". Cancer Research. 70 (19): 7500–13. doi:10.1158/0008-5472.CAN-10-2353. PMID 20858720.
- ↑ Anido, J; Sáez-Borderías, A; Gonzàlez-Juncà, A; Rodón, L; Folch, G; Carmona, MA; Prieto-Sánchez, RM; Barba, I; Martínez-Sáez, E; Prudkin, L; Cuartas, I; Raventós, C; Martínez-Ricarte, F; Poca, MA; García-Dorado, D; Lahn, MM; Yingling, JM; Rodón, J; Sahuquillo, J; Baselga, J; Seoane, J (Dec 14, 2010). "TGF-β Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma". Cancer Cell. 18 (6): 655–68. doi:10.1016/j.ccr.2010.10.023. PMID 21156287.
- 1 2 O'Brien, CA; Pollett, A; Gallinger, S; Dick, JE (Jan 4, 2007). "A human colon cancer cell capable of initiating tumour growth in immunodeficient mice". Nature. 445 (7123): 106–10. doi:10.1038/nature05372. PMID 17122772.
- 1 2 Dalerba, P; Dylla, SJ; Park, IK; Liu, R; Wang, X; Cho, RW; Hoey, T; Gurney, A; Huang, EH; Simeone, DM; Shelton, AA; Parmiani, G; Castelli, C; Clarke, MF (Jun 12, 2007). "Phenotypic characterization of human colorectal cancer stem cells". Proceedings of the National Academy of Sciences of the United States of America. 104 (24): 10158–63. doi:10.1073/pnas.0703478104. PMC 1891215
 . PMID 17548814.
. PMID 17548814. - ↑ Wilson, BJ; Schatton, T; Zhan, Q; Gasser, M; Ma, J; Saab, KR; Schanche, R; Waaga-Gasser, AM; Gold, JS; Huang, Q; Murphy, GF; Frank, MH; Frank, NY (Aug 1, 2011). "ABCB5 identifies a therapy-refractory tumor cell population in colorectal cancer patients". Cancer Research. 71 (15): 5307–16. doi:10.1158/0008-5472.CAN-11-0221. PMC 3395026
 . PMID 21652540.
. PMID 21652540. - ↑ Odoux, C; Fohrer, H; Hoppo, T; Guzik, L; Stolz, DB; Lewis, DW; Gollin, SM; Gamblin, TC; Geller, DA; Lagasse, E (Sep 1, 2008). "A stochastic model for cancer stem cell origin in metastatic colon cancer". Cancer Research. 68 (17): 6932–41. doi:10.1158/0008-5472.CAN-07-5779. PMC 2562348
 . PMID 18757407.
. PMID 18757407. - ↑ Huang, EH; Hynes, MJ; Zhang, T; Ginestier, C; Dontu, G; Appelman, H; Fields, JZ; Wicha, MS; Boman, BM (Apr 15, 2009). "Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis". Cancer Research. 69 (8): 3382–9. doi:10.1158/0008-5472.CAN-08-4418. PMC 2789401
 . PMID 19336570.
. PMID 19336570. - ↑ Kemper, K; Sprick, MR; de Bree, M; Scopelliti, A; Vermeulen, L; Hoek, M; Zeilstra, J; Pals, ST; Mehmet, H; Stassi, G; Medema, JP (Jan 15, 2010). "The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation". Cancer Research. 70 (2): 719–29. doi:10.1158/0008-5472.CAN-09-1820. PMID 20068153.
- ↑ Liu, C; Kelnar, K; Liu, B; Chen, X; Calhoun-Davis, T; Li, H; Patrawala, L; Yan, H; Jeter, C; Honorio, S; Wiggins, JF; Bader, AG; Fagin, R; Brown, D; Tang, DG (February 2011). "The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44". Nature Medicine. 17 (2): 211–5. doi:10.1038/nm.2284. PMC 3076220
 . PMID 21240262.
. PMID 21240262. - ↑ Ho, MM; Ng, AV; Lam, S; Hung, JY (May 15, 2007). "Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells". Cancer Research. 67 (10): 4827–33. doi:10.1158/0008-5472.CAN-06-3557. PMID 17510412.
- ↑ Patrawala, L; Calhoun, T; Schneider-Broussard, R; Li, H; Bhatia, B; Tang, S; Reilly, JG; Chandra, D; Zhou, J; Claypool, K; Coghlan, L; Tang, DG (Mar 16, 2006). "Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells". Oncogene. 25 (12): 1696–708. doi:10.1038/sj.onc.1209327. PMID 16449977.
- ↑ Dubrovska, A; Kim, S; Salamone, RJ; Walker, JR; Maira, SM; García-Echeverría, C; Schultz, PG; Reddy, VA (Jan 6, 2009). "The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations". Proceedings of the National Academy of Sciences of the United States of America. 106 (1): 268–73. doi:10.1073/pnas.0810956106. PMC 2629188
 . PMID 19116269.
. PMID 19116269. - ↑ Rajasekhar, VK; Studer, L; Gerald, W; Socci, ND; Scher, HI (Jan 18, 2011). "Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-κB signalling". Nature Communications. 2 (1): 162–. doi:10.1038/ncomms1159. PMC 3105310
 . PMID 21245843.
. PMID 21245843. - ↑ Li, T; Su, Y; Mei, Y; Leng, Q; Leng, B; Liu, Z; Stass, SA; Jiang, F (February 2010). "ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome". Laboratory Investigation. 90 (2): 234–44. doi:10.1038/labinvest.2009.127. PMC 3552330
 . PMID 20010854.
. PMID 20010854. - ↑ Eramo, A; Lotti, F; Sette, G; Pilozzi, E; Biffoni, M; Di Virgilio, A; Conticello, C; Ruco, L; Peschle, C; De Maria, R (March 2008). "Identification and expansion of the tumorigenic lung cancer stem cell population". Cell death and differentiation. 15 (3): 504–14. doi:10.1038/sj.cdd.4402283. PMID 18049477.
- ↑ Sullivan, JP; Spinola, M; Dodge, M; Raso, MG; Behrens, C; Gao, B; Schuster, K; Shao, C; Larsen, JE; Sullivan, LA; Honorio, S; Xie, Y; Scaglioni, PP; DiMaio, JM; Gazdar, AF; Shay, JW; Wistuba, II; Minna, JD (Dec 1, 2010). "Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling". Cancer Research. 70 (23): 9937–48. doi:10.1158/0008-5472.CAN-10-0881. PMC 3058307
 . PMID 21118965.
. PMID 21118965. - ↑ Leung, EL; Fiscus, RR; Tung, JW; Tin, VP; Cheng, LC; Sihoe, AD; Fink, LM; Ma, Y; Wong, MP (Nov 19, 2010). Jin, Dong-Yan, ed. "Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties". PLOS ONE. 5 (11): e14062. doi:10.1371/journal.pone.0014062. PMC 2988826
 . PMID 21124918.
. PMID 21124918. - ↑ Damelin, M; Geles, KG; Follettie, MT; Yuan, P; Baxter, M; Golas, J; DiJoseph, JF; Karnoub, M; Huang, S; Diesl, V; Behrens, C; Choe, SE; Rios, C; Gruzas, J; Sridharan, L; Dougher, M; Kunz, A; Hamann, PR; Evans, D; Armellino, D; Khandke, K; Marquette, K; Tchistiakova, L; Boghaert, ER; Abraham, RT; Wistuba, II; Zhou, BB (Jun 15, 2011). "Delineation of a cellular hierarchy in lung cancer reveals an oncofetal antigen expressed on tumor-initiating cells". Cancer Research. 71 (12): 4236–46. doi:10.1158/0008-5472.CAN-10-3919. PMID 21540235.
- ↑ Thiery, JP (June 2002). "Epithelial-mesenchymal transitions in tumour progression". Nature Reviews Cancer. 2 (6): 442–54. doi:10.1038/nrc822. PMID 12189386.
- ↑ Angerer, LM; Angerer, RC (June 1999). "Regulative development of the sea urchin embryo: signalling cascades and morphogen gradients". Seminars in cell & developmental biology. 10 (3): 327–34. doi:10.1006/scdb.1999.0292. PMID 10441547.
- ↑ Mani, SA; Yang, J; Brooks, M; Schwaninger, G; Zhou, A; Miura, N; Kutok, JL; Hartwell, K; Richardson, AL; Weinberg, RA (Jun 12, 2007). "Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers". Proceedings of the National Academy of Sciences of the United States of America. 104 (24): 10069–74. doi:10.1073/pnas.0703900104. PMC 1891217
 . PMID 17537911.
. PMID 17537911. - ↑ Conacci-Sorrell, M; Simcha, I; Ben-Yedidia, T; Blechman, J; Savagner, P; Ben-Ze'ev, A (Nov 24, 2003). "Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK". The Journal of Cell Biology. 163 (4): 847–57. doi:10.1083/jcb.200308162. PMC 2173691
 . PMID 14623871.
. PMID 14623871. - ↑ Spill, F.; Reynolds, D.S.; Kamm, R.D.; Zaman, M.H. "Impact of the physical microenvironment on tumor progression and metastasis". Current Opinion in Biotechnology. 40: 41–48. doi:10.1016/j.copbio.2016.02.007.
- ↑ Kaplan, RN; Riba, RD; Zacharoulis, S; Bramley, AH; Vincent, L; Costa, C; MacDonald, DD; Jin, DK; Shido, K; Kerns, SA; Zhu, Z; Hicklin, D; Wu, Y; Port, JL; Altorki, N; Port, ER; Ruggero, D; Shmelkov, SV; Jensen, KK; Rafii, S; Lyden, D (Dec 8, 2005). "VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche". Nature. 438 (7069): 820–7. doi:10.1038/nature04186. PMC 2945882
 . PMID 16341007.
. PMID 16341007. - ↑ Hermann, PC; Huber, SL; Herrler, T; Aicher, A; Ellwart, JW; Guba, M; Bruns, CJ; Heeschen, C (Sep 13, 2007). "Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer". Cell stem cell. 1 (3): 313–23. doi:10.1016/j.stem.2007.06.002. PMID 18371365.
- ↑ Yang, ZF; Ho, DW; Ng, MN; Lau, CK; Yu, WC; Ngai, P; Chu, PW; Lam, CT; Poon, RT; Fan, ST (February 2008). "Significance of CD90+ cancer stem cells in human liver cancer". Cancer Cell. 13 (2): 153–66. doi:10.1016/j.ccr.2008.01.013. PMID 18242515.
- ↑ Shipitsin, M; Campbell, LL; Argani, P; Weremowicz, S; Bloushtain-Qimron, N; Yao, J; Nikolskaya, T; Serebryiskaya, T; Beroukhim, R; Hu, M; Halushka, MK; Sukumar, S; Parker, LM; Anderson, KS; Harris, LN; Garber, JE; Richardson, AL; Schnitt, SJ; Nikolsky, Y; Gelman, RS; Polyak, K (March 2007). "Molecular definition of breast tumor heterogeneity". Cancer Cell. 11 (3): 259–73. doi:10.1016/j.ccr.2007.01.013. PMID 17349583.
- ↑ Shmelkov, SV; Butler, JM; Hooper, AT; Hormigo, A; Kushner, J; Milde, T; St Clair, R; Baljevic, M; White, I; Jin, DK; Chadburn, A; Murphy, AJ; Valenzuela, DM; Gale, NW; Thurston, G; Yancopoulos, GD; D'Angelica, M; Kemeny, N; Lyden, D; Rafii, S (June 2008). "CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors". The Journal of Clinical Investigation. 118 (6): 2111–20. doi:10.1172/JCI34401. PMC 2391278
 . PMID 18497886.
. PMID 18497886. - ↑ Brabletz, T; Jung, A; Spaderna, S; Hlubek, F; Kirchner, T (September 2005). "Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression". Nature Reviews Cancer. 5 (9): 744–9. doi:10.1038/nrc1694. PMID 16148886.
- 1 2 Mraz, M.; Zent, C. S.; Church, A. K.; Jelinek, D. F.; Wu, X.; Pospisilova, S.; Ansell, S. M.; Novak, A. J.; Kay, N. E.; Witzig, T. E.; Nowakowski, G. S. (2011). "Bone marrow stromal cells protect lymphoma B-cells from rituximab-induced apoptosis and targeting integrin α-4-β-1 (VLA-4) with natalizumab can overcome this resistance". British Journal of Haematology. 155 (1): 53–64. doi:10.1111/j.1365-2141.2011.08794.x. PMID 21749361.
- ↑ Dirks, P (Jul 1, 2010). "Cancer stem cells: Invitation to a second round". Nature. 466 (7302): 40–1. doi:10.1038/466040a. PMID 20596007.
- ↑ Haskins WE, Eedala S, Jadhav YL, Labhan MS, Pericherla VC, Perlman EJ (2012). "Insights on neoplastic stem cells from gel-based proteomics of childhood germ cell tumors". Pediatr Blood Cancer. 58: 722–8. doi:10.1002/pbc.23282. PMC 3204330
 . PMID 21793190.
. PMID 21793190. - ↑ Gupta, PB; Onder, TT; Jiang, G; Tao, K; Kuperwasser, C; Weinberg, RA; Lander, ES (Aug 21, 2009). "Identification of selective inhibitors of cancer stem cells by high-throughput screening". Cell. 138 (4): 645–59. doi:10.1016/j.cell.2009.06.034. PMID 19682730.
- ↑ Jangamreddy JR, Ghavami S, Grabarek J, Kratz G, Wiechec E, Fredriksson BA, Rao Pariti RK, Cieślar-Pobuda A, Panigrahi S, Łos MJ (2013). "Salinomycin induces activation of autophagy, mitophagy and affects mitochondrial polarity: differences between primary and cancer cells.". Biochim Biophys Acta. 1833 (9): 2057–69. doi:10.1016/j.bbamcr.2013.04.011. PMID 23639289.
- ↑ Vlahopoulos S, Critselis E, Voutsas IF, Perez SA, Moschovi M, Baxevanis CN, Chrousos GP (2014). "New use for old drugs? Prospective targets of chloroquines in cancer therapy". Curr Drug Targets. 15 (9): 843–51. doi:10.2174/1389450115666140714121514. PMID 25023646.
- ↑ Jordan, C. T.; Upchurch, D.; Szilvassy, S. J.; Guzman, M. L.; Howard, D. S.; Pettigrew, A. L.; Meyerrose, T.; Rossi, R.; Grimes, B. (2000-10-01). "The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells". Leukemia. 14 (10): 1777–1784. doi:10.1038/sj.leu.2401903. ISSN 0887-6924. PMID 11021753.
- ↑ Jin, Liqing; Lee, Erwin M.; Ramshaw, Hayley S.; Busfield, Samantha J.; Peoppl, Armando G.; Wilkinson, Lucy; Guthridge, Mark A.; Thomas, Daniel; Barry, Emma F. (2009-07-02). "Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells". Cell Stem Cell. 5 (1): 31–42. doi:10.1016/j.stem.2009.04.018. ISSN 1875-9777. PMID 19570512.
- ↑ "'Nanobombs' that blow up cancer cells | KurzweilAI". www.kurzweilai.net. 2015-12-07. Retrieved 2016-02-20.
- ↑ Wang, Hai; Agarwal, Pranay; Zhao, Shuting; Yu, Jianhua; Lu, Xiongbin; He, Xiaoming (2016-01-01). "A Near-Infrared Laser-Activated "Nanobomb" for Breaking the Barriers to MicroRNA Delivery". Advanced Materials. 28 (2): 347–355. doi:10.1002/adma.201504263. ISSN 1521-4095.
- ↑ Haupt Y, Bath ML, Harris AW, Adams JM (November 1993). "bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis". Oncogene. 8 (11): 3161–4. PMID 8414519.
- ↑ Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (May 2003). "Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells". Nature. 423 (6937): 302–5. doi:10.1038/nature01587. PMID 12714971.
- ↑ Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ (October 2003). "Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation". Nature. 425 (6961): 962–7. doi:10.1038/nature02060. PMC 2614897
 . PMID 14574365.
. PMID 14574365. - ↑ Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI (December 2003). "Cancerous stem cells can arise from pediatric brain tumors". Proceedings of the National Academy of Sciences of the United States of America. 100 (25): 15178–83. doi:10.1073/pnas.2036535100. PMC 299944
 . PMID 14645703.
. PMID 14645703. - ↑ Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS (2004). "Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells". Breast cancer research : BCR. 6 (6): R605–15. doi:10.1186/bcr920. PMC 1064073
 . PMID 15535842.
. PMID 15535842. - ↑ Diévart A, Beaulieu N, Jolicoeur P (October 1999). "Involvement of Notch1 in the development of mouse mammary tumors". Oncogene. 18 (44): 5973–81. doi:10.1038/sj.onc.1202991. PMID 10557086.
- ↑ Park DM, Jung J, Masjkur J, et al. (2013). "Hes3 regulates cell number in cultures from glioblastoma multiforme with stem cell characteristics". Sci Rep. 3: 1095. doi:10.1038/srep01095. PMC 3566603
 . PMID 23393614.
. PMID 23393614. - ↑ Beachy PA, Karhadkar SS, Berman DM (November 2004). "Tissue repair and stem cell renewal in carcinogenesis". Nature. 432 (7015): 324–31. doi:10.1038/nature03100. PMID 15549094.
- ↑ Zhou BP, Hung MC (June 2005). "Wnt, hedgehog and snail: sister pathways that control by GSK-3beta and beta-Trcp in the regulation of metastasis". Cell cycle (Georgetown, Tex.). 4 (6): 772–6. doi:10.4161/cc.4.6.1744. PMID 15917668.
- ↑ Akiyoshi T, Nakamura M, Koga K, Nakashima H, Yao T, Tsuneyoshi M, Tanaka M, Katano M (July 2006). "Gli1, downregulated in colorectal cancers, inhibits proliferation of colon cancer cells involving Wnt signalling activation". Gut. 55 (7): 991–9. doi:10.1136/gut.2005.080333. PMC 1856354
 . PMID 16299030.
. PMID 16299030. - ↑ She M, Chen X (2009). "Targeting signal pathways active in cancer stem cells to overcome drug resistance". Chin J Lung Cancer. 12 (1): 3–7. doi:10.3779/j.issn.1009-3419.2009.01.001. PMID 20712949.
- ↑ Bollmann FM (August 2008). "The many faces of telomerase: emerging extratelomeric effects". BioEssays. 30 (8): 728–32. doi:10.1002/bies.20793. PMID 18623070.
- ↑ Hodge, Russ (2016-01-25). "Hacking the programs of cancer stem cells". medicalxpress.com. Medical Express. Retrieved 2016-02-12.
Further reading
- Polyak K, Weinberg RA (April 2009). "Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits". Nature Reviews Cancer. 9 (4): 265–73. doi:10.1038/nrc2620. PMID 19262571.
- Sánchez-García I, Vicente-Dueñas C, Cobaleda C (December 2007). "The theoretical basis of cancer-stem-cell-based therapeutics of cancer: can it be put into practice?". BioEssays. 29 (12): 1269–80. doi:10.1002/bies.20679. PMID 18022789.
- Gao JX (2008). "Cancer stem cells: the lessons from pre-cancerous stem cells". Journal of Cellular and Molecular Medicine. 12 (1): 67–96. doi:10.1111/j.1582-4934.2007.00170.x. PMID 18053092.
- Yanyan Lia; Tao Zhang (May 2014). "Targeting cancer stem cells by curcumin and clinical applications". Cancer Letters. 346 (2): 197–205. doi:10.1016/j.canlet.2014.01.012.
External links
| Wikimedia Commons has media related to Cancer stem cells. |
- European Cancer Stem Cell Research Institute A new Institute dedicated to research into cancer stem cells and related work.
- Cancer Stem Cell News A blog of news items related to cancer stem cells, with an emphasis on recent research and articles that are openly accessible
- Exploring the role of cancer stem cells in radioresistance Abstract of a review by Michael Baumann, Mechthild Krause, Richard Hill, "Nature Reviews Cancer " 2008(Jul); 8(7) 545-54
- "A Tumor's Lifeblood", Jessica Gorman, CR magazine, Summer 2006
- "Cancer Stem Cell Scientific Literature Review", UMDNJ Stem Cell Research and Regenerative Medicine, June 17, 2006
- "Stem cells may cause some forms of bone cancer", News-Medical.Net, December 7, 2005
- "The Bad Seed: Rare stem cells appear to drive cancers", Science News Online, March 20, 2004
- "The Real Problem in Breast Tumors: Cancer Stem Cells", Genome News Network, March 7, 2003
- Differentiation Therapy - A Different Approach to Treating Tumors (from Beaker Blog)
- Characteristics of Cancer Cells Cancer Inform Blog
- Cancer stem cells may be cause of brain tumors (research of John A. Boockvar)