Cadherin
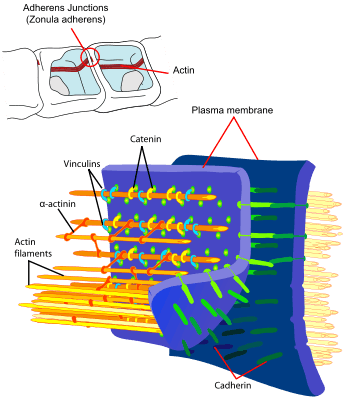
Cadherins (named for "calcium-dependent adhesion") are a class of type-1 transmembrane proteins. They play important roles in cell adhesion, forming adherens junctions to bind cells within tissues together. They are dependent on calcium (Ca2+) ions to function, hence their name. Cell-cell adhesion is mediated by extracellular cadherin domains, whereas the intracellular cytoplasmic tail associates with a large number of adaptor and signaling proteins, collectively referred to as the cadherin adhesome.
The cadherin superfamily includes cadherins, protocadherins, desmogleins, and desmocollins, and more.[1][2] In structure, they share cadherin repeats, which are the extracellular Ca2+-binding domains. There are multiple classes of cadherin molecule, each designated with a prefix (in general, noting the type of tissue with which it is associated). It has been observed that cells containing a specific cadherin subtype tend to cluster together to the exclusion of other types, both in cell culture and during development.[3] For example, cells containing N-cadherin tend to cluster with other N-cadherin-expressing cells. However, it has been noted that the mixing speed in the cell culture experiments can have an effect on the extent of homotypic specificity.[4] In addition, several groups have observed heterotypic binding affinity (i.e., binding of different types of cadherin together) in various assays.[5][6] One current model proposes that cells distinguish cadherin subtypes based on kinetic specificity rather than thermodynamic specificity, as different types of cadherin homotypic bonds have different lifetimes.[7]
Structure and Function
Cadherins are synthesized as polypeptides and undergo many post-translational modifications to become the proteins which mediate cell-cell adhesion and recognition.[8] These polypeptides are approximately 720–750 amino acids long. Each cadherin has a small cytoplasmic component, a transmembrane component, and the remaining bulk of the protein is extra-cellular (outside the cell). To date, over 100 types of cadherins in humans have been identified and sequenced.[9]
Development
Cadherins behave as both receptors and ligands for other molecules. During development, their behavior assists in properly positioning cells: they are responsible for the separation of the different tissue layers, and for cellular migration.[10] In the very early stages of development, E-cadherin (epithelial cadherin) is most greatly expressed. During the next stage, the development of the neural plate, N-cadherin (neural cadherin) is expressed and there is a decrease in E-cadherin. Finally, during the development of the notochord and the condensation of somites, E- P- and N-cadherin expression increases. After development, cadherins play a role in maintaining cell and tissue structure, and in cellular movement.[9] Regulation of cadherin expression can occur through promoter methylation among other epigenetic mechanisms.[11]
Tumour metastasis
The E-cadherin - catenin complex plays a key role in cellular adhesion; loss of this function has been associated with greater tumour metastasis.[12]
Types
| Cadherin domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Cadherin | ||||||||
| Pfam | PF00028 | ||||||||
| InterPro | IPR002126 | ||||||||
| SMART | CA | ||||||||
| PROSITE | PDOC00205 | ||||||||
| SCOP | 1nci | ||||||||
| SUPERFAMILY | 1nci | ||||||||
| |||||||||

There are said to be over 100 different types of cadherins found in vertebrates, which can be classified into four groups: classical, desmosomal, protocadherins, and unconventional.[14][15] This large amount of diversity is accomplished by having multiple cadherin encoding genes combined with alternative RNA splicing mechanisms. Invertebrates contain fewer than 20 types of cadherins.[15]
Classical
Different members of the cadherin family are found in different locations.
- CDH1 - E-cadherin (epithelial): E-cadherins are found in epithelial tissue
- CDH2 - N-cadherin (neural): N-cadherins are found in neurons
- CDH12 - cadherin 12, type 2 (N-cadherin 2)
- CDH3 - P-cadherin (placental): P-cadherins are found in the placenta.
Desmosomal
- Desmoglein (DSG1, DSG2, DSG3, DSG4)
- Desmocollin (DSC1, DSC2, DSC3)
Protocadherins
PCDH15; PCDH17; PCDH18; PCDH19; PCDH20; PCDH7; PCDH8; PCDH9; PCDHA1; PCDHA10; PCDHA11; PCDHA12; PCDHA13; PCDHA2; PCDHA3; PCDHA4; PCDHA5; PCDHA6; PCDHA7; PCDHA8; PCDHA9; PCDHAC1; PCDHAC2; PCDHB1; PCDHB10; PCDHB11; PCDHB12; PCDHB13; PCDHB14; PCDHB15; PCDHB16; PCDHB17; PCDHB18; PCDHB2; PCDHB3; PCDHB4; PCDHB5; PCDHB6; PCDHB7; PCDHB8; PCDHB9; PCDHGA1; PCDHGA10; PCDHGA11; PCDHGA12; PCDHGA2; PCDHGA3; PCDHGA4; PCDHGA5; PCDHGA6; PCDHGA7; PCDHGA8; PCDHGA9; PCDHGB1; PCDHGB2; PCDHGB3; PCDHGB4; PCDHGB5; PCDHGB6; PCDHGB7; PCDHGC3; PCDHGC4; PCDHGC5
Unconventional/ungrouped
- CDH9 - cadherin 9, type 2 (T1-cadherin)
- CDH10 - cadherin 10, type 2 (T2-cadherin)
- CDH4 - R-cadherin (retinal)
- CDH5 - VE-cadherin (vascular endothelial)
- CDH6 - K-cadherin (kidney)
- CDH7 - cadherin 7, type 2
- CDH8 - cadherin 8, type 2
- CDH11 - OB-cadherin (osteoblast)
- CDH13 - T-cadherin - H-cadherin (heart)
- CDH15 - M-cadherin (myotubule)
- CDH16 - KSP-cadherin
- CDH17 - LI cadherin (liver-intestine)
- CDH18 - cadherin 18, type 2
- CDH19 - cadherin 19, type 2
- CDH20 - cadherin 20, type 2
- CDH23 - cadherin 23 (neurosensory epithelium)
- CDH10; CDH11; CDH13; CDH15; CDH16; CDH17;
CDH18; CDH19; CDH20; CDH22; CDH23; CDH24; CDH26; CDH28; CDH4; CDH5; CDH6; CDH7; CDH8; CDH9;
CELSR1; CELSR2; CELSR3; CLSTN1; CLSTN2; CLSTN3; DCHS1; DCHS2; LOC389118;
See also
References
- ↑ Hulpiau P, van Roy F (February 2009). "Molecular evolution of the cadherin superfamily". Int. J. Biochem. Cell Biol. 41 (2): 349–69. doi:10.1016/j.biocel.2008.09.027. PMID 18848899.
- ↑ Angst B, Marcozzi C, Magee A (February 2001). "The cadherin superfamily: diversity in form and function". J Cell Sci. 114 (Pt 4): 629–41. PMID 11171368.
- ↑ Bello, S.M.; Millo, H.; Rajebhosale, M.; Price, S.R. (2012). "Catenin-dependent cadherin function drives divisional segregation of spinal chord motor neurons". J. Neuroscience. 32 (2): 490–505. doi:10.1523/jneurosci.4382-11.2012.
- ↑ Duguay, D.; A. Foty R., RA; S. Steinberg M., MS (2003). "Cadherin-mediated cell adhesion and tissue segregation: qualitative and quantitative determinants". Dev. Biol. 253 (2): 309–323. doi:10.1016/S0012-1606(02)00016-7. PMID 12645933.
- ↑ Niessen, Carien M.; Gumbiner, Barry M. (2002). "Cadherin-mediated cell sorting not determined by binding or adhesion specificity". The Journal of Cell Biology. 156 (2): 389–399. doi:10.1083/jcb.200108040. PMC 2199232
 . PMID 11790800.
. PMID 11790800. - ↑ Volk, T.; Cohen, O.; Geiger, B. (1987). "Formation of heterotypic adherens-type junctions between L-CAM containing liver cells and A-CAM containing lens cells". Cell. 50 (6): 987–994. doi:10.1016/0092-8674(87)90525-3. PMID 3621349.
- ↑ Bayas, Marco V.; Leung, Andrew; Evans, Evan; Leckband, Deborah (2005). "Lifetime Measurements Reveal Kinetic Differences between Homophilic Cadherin Bonds". Biophysical Journal. 90 (4): 1385–95. doi:10.1529/biophysj.105.069583. PMC 1367289
 . PMID 16326909.
. PMID 16326909. - ↑ Harris, T. J.; Tepass, U (2010). "Adherens junctions: From molecules to morphogenesis". Nature Reviews Molecular Cell Biology. 11 (7): 502–14. doi:10.1038/nrm2927. PMID 20571587.
- 1 2 Tepass, U; Truong, K; Godt, D; Ikura, M; Peifer, M (2000). "Cadherins in embryonic and neural morphogenesis". Nature Reviews Molecular Cell Biology. 1 (2): 91–100. doi:10.1038/35040042. PMID 11253370.
- ↑ Gumbiner, B. M. (2005). "Regulation of cadherin-mediated adhesion in morphogenesis". Nature Reviews Molecular Cell Biology. 6 (8): 622–34. doi:10.1038/nrm1699. PMID 16025097.
- ↑ Reinhold, WC; Reimers, MA; Maunakea, AK; Kim, S; Lababidi, S; Scherf, U; Shankavaram, UT; Ziegler, MS; Stewart, C; Kouros-Mehr, Hosein; Cui, H; Dolginow, D; Scudiero, DA; Pommier, YG; Munroe, DJ; Feinberg, AP; Weinstein, JN (Feb 2007). "Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells.". Molecular cancer therapeutics. 6 (2): 391–403. doi:10.1158/1535-7163.MCT-06-0609. PMID 17272646.
- ↑ Beavon, IR (August 2000). "The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation.". European journal of cancer (Oxford, England : 1990). 36 (13 Spec No): 1607–20. doi:10.1016/S0959-8049(00)00158-1. PMID 10959047.
- ↑ PDB: 3Q2V; Harrison, O.J.; Jin, X.; Hong, S.; Bahna, F.; Ahlsen, G.; Brasch, J.; Wu, Y.; Vendome, J.; Felsovalyi, K.; Hampton, C.M.; Troyanovsky, R.B.; Ben-Shaul, A.; Frank, J.; Troyanovsky, S.M.; Shapiro, L.; Honig, B. (2011). "The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins". Structure. 19 (2): 244–56. doi:10.1016/j.str.2010.11.016. PMC 3070544
 . PMID 21300292.; rendered with PyMOL
. PMID 21300292.; rendered with PyMOL - ↑ Stefan Offermanns; Walter Rosenthal (2008). Encyclopedia of Molecular Pharmacology. Springer. pp. 306–. ISBN 978-3-540-38916-3. Retrieved 14 December 2010.
- 1 2 Lodish, Harvey; Berk, Arnold; Kaiser, Chris; Krieger, Monte; Bretscher, Anthony; Ploegh, Hidde; Amon, Angelika (2013). Molecular Cell Biology (Seventh ed.). New York: Worth Publ. p. 934. ISBN 978-1-4292-3413-9.
Further reading
- Beavon IR (2000). "The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation". Eur. J. Cancer. 36 (13 Spec No): 1607–20. doi:10.1016/S0959-8049(00)00158-1. PMID 10959047.
- Berx G, Becker KF, Höfler H, van Roy F (1998). "Mutations of the human E-cadherin (CDH1) gene". Hum. Mutat. 12 (4): 226–37. doi:10.1002/(SICI)1098-1004(1998)12:4<226::AID-HUMU2>3.0.CO;2-D. PMID 9744472.
- Bryant DM, Stow JL (2005). "The ins and outs of E-cadherin trafficking". Trends Cell Biol. 14 (8): 427–34. doi:10.1016/j.tcb.2004.07.007. PMID 15308209.
- Chun YS, Lindor NM, Smyrk TC, et al. (2001). "Germline E-cadherin gene mutations: is prophylactic total gastrectomy indicated?". Cancer. 92 (1): 181–7. doi:10.1002/1097-0142(20010701)92:1<181::AID-CNCR1307>3.0.CO;2-J. PMID 11443625.
- Georgolios A, Batistatou A, Manolopoulos L, Charalabopoulos K (2006). "Role and expression patterns of E-cadherin in head and neck squamous cell carcinoma (HNSCC)". J. Exp. Clin. Cancer Res. 25 (1): 5–14. PMID 16761612.
- Hazan RB, Qiao R, Keren R, et al. (2004). "Cadherin switch in tumor progression". Ann. N. Y. Acad. Sci. 1014 (1): 155–63. doi:10.1196/annals.1294.016. PMID 15153430.
- Moran CJ, Joyce M, McAnena OJ (2005). "CDH1 associated gastric cancer: a report of a family and review of the literature". Eur J Surg Oncol. 31 (3): 259–64. doi:10.1016/j.ejso.2004.12.010. PMID 15780560.
- Reynolds AB, Carnahan RH (2005). "Regulation of cadherin stability and turnover by p120ctn: implications in disease and cancer". Semin. Cell Dev. Biol. 15 (6): 657–63. doi:10.1016/j.semcdb.2004.09.003. PMID 15561585.
- Wang HD, Ren J, Zhang L (2004). "CDH1 germline mutation in hereditary gastric carcinoma". World J. Gastroenterol. 10 (21): 3088–93. PMID 15457549.
- Wijnhoven BP, Dinjens WN, Pignatelli M (2000). "E-cadherin-catenin cell-cell adhesion complex and human cancer". The British journal of surgery. 87 (8): 992–1005. doi:10.1046/j.1365-2168.2000.01513.x. PMID 10931041.
- Wilson PD (2001). "Polycystin: new aspects of structure, function, and regulation". J. Am. Soc. Nephrol. 12 (4): 834–45. PMID 11274246.
- Renaud-Young M, Gallin WJ (2002). "In the first extracellular domain of E-cadherin, heterophilic interactions, but not the conserved His-Ala-Val motif, are required for adhesion". Journal of Biological Chemistry. 277 (42): 39609–39616. doi:10.1074/jbc.M201256200. PMID 12154084.
External links
- Proteopedia Cadherin - view cadherin structure in interactive 3D
- Cadherin domain in PROSITE
- The cadherin family
- Alberts, Bruce. Molecular Biology of the Cell
- The Cadherin Resource
- InterPro: IPR002126
- "Cadherin adhesome at a glance". J Cell Sci 126, 373-378