Eukaryotic translation
Eukaryotic translation is the process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recycling.
Initiation
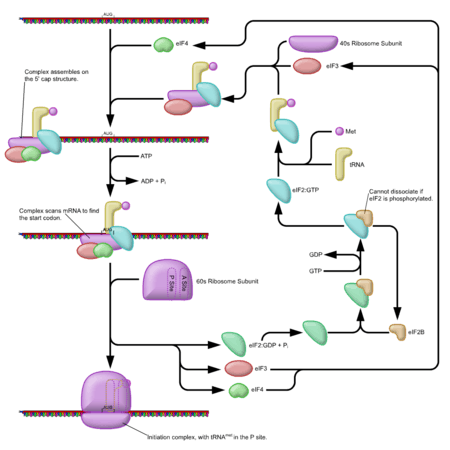
Cap-dependent initiation
Initiation of translation usually involves the interaction of certain key proteins with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap, as well as with the 5' UTR. The protein factors bind the small ribosomal subunit (also referred to as the 40S subunit), and these initiation factors hold the mRNA in place. eIF3 is associated with the 40S ribosomal subunit, and plays a role in keeping the large (60S) ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex, which consists of three other initiation factors: eIF4A, eIF4E, and eIF4G. eIF4G is a scaffolding protein that directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. Binding of the cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control. Certain viruses cleave a portion of eIF4G that binds eIF4E, thus preventing cap-dependent translation to hijack the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase, which aids the ribosome in resolving certain secondary structures formed along the mRNA transcript.[1]
The Poly(A)-binding protein (PABP) also associates with the eIF4F complex via eIF4G, and binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.[2][3] This 43S pre-initiation complex (PIC) accompanied by the protein factors moves along the mRNA chain toward its 3'-end, in a process known as 'scanning', to reach the start codon (typically AUG). In eukaryotes and archaea, the amino acid encoded by the start codon is methionine. The Met-charged initiator tRNA (Met-tRNAiMet) is brought to the P-site of the small ribosomal subunit by eukaryotic initiation factor 2 (eIF2). It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit, eventually leading to the association of the large subunit (or the 60S subunit). The complete ribosome (80S) then commences translation elongation.
Regulation of protein synthesis partly influenced by phosphorylation of eIF2 (via the α subunit), which is a part of the eIF2-GTP-Met-tRNAiMet ternary complex (TC). When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This occurs if there is amino acid starvation or there has been a virus infection. However, naturally a small percentage of this initiation factor is phosphorylated. Another regulator is 4EBP, which binds to the initiation factor eIF4E and inhibits its interactions with eIF4G, thus preventing cap-dependent initiation. To oppose the effects of the 4EBP, growth factors phosphorylate 4EBP, reducing its affinity for eIF4E and permitting protein synthesis.
While the global regulation of protein synthesis is achieved by modulating the expression of key initiation factors as well as the number of ribosomes, individual mRNAs can have different translation rates due to presence of regulatory sequence elements. This has been shown to be important in a variety of settings including yeast meiosis and ethylene response in plants. In addition, recent work in yeast and humans suggest that evolutionary divergence in cis-regulatory sequences can impact translation regulation.[4]
The cap-independent initiation
The best-studied example of cap-independent translation initiation in eukaryotes is that by the Internal ribosome entry site (IRES). What differentiates cap-independent translation from cap-dependent translation is that cap-independent translation does not require the 5' cap to initiate scanning from the 5' end of the mRNA until the start codon. The ribosome can be trafficked to the start site by direct binding, initiation factors, and/or ITAFs (IRES trans-acting factors) bypassing the need to scan the entire 5' UTR. This method of translation has been found important in conditions that require the translation of specific mRNAs during cellular stress, when overall translation is reduced. Examples include factors responding to apoptosis and stress-induced responses.[5]
Elongation

Elongation depends on eukaryotic elongation factors. At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, (2) forming the peptide bond and (3) shifting the mRNA by one codon relative to the ribosome.
Unlike bacteria, in which translation initiation occurs as soon as the 5' end of an mRNA is synthesized, in eukaryotes such tight coupling between transcription and translation is not possible because transcription and translation are carried out in separate compartments of the cell (the nucleus and cytoplasm). Eukaryotic mRNA precursors must be processed in the nucleus (e.g., capping, polyadenylation, splicing) before they are exported to the cytoplasm for translation.
Translation can also be affected by ribosomal pausing, which can trigger endonucleolytic attack of the mRNA, a process termed mRNA no-go decay. Ribosomal pausing also aids co-translational folding of the nascent polypeptide on the ribosome, and delays protein translation while it is encoding mRNA. This can trigger ribosomal frameshifting.[6]
Termination
Termination of elongation depends on eukaryotic release factors. The process is similar to that of prokaryotic termination, but unlike prokaryotic termination, there is a universal release factor, eRF1, that recognizes all three stop codons. Upon Termination, the ribosome is disassembled and the completed polypeptide is released. eRF3 is a ribosome-dependent GTPase that helps eRF1 release the completed polypeptide. The human genome encodes a few genes, whose mRNA stop codon are surprisingly leaky: In these genes, termination of translation is inefficient due to special RNA bases in the vicinity of the stoo codon. Leaky termination in these genes leads to translational readthrough of up to 10% of the stop codons of these genes. Some of these genes encode functional protein domains in their readthrough extension so that new protein isoforms can arise. This process has been termed 'functional tranlational readthrough'. [7]
See also
- 40S
- 60S
- 80S
- Eukaryotic initiation factor
- Eukaryotic elongation factors
- Eukaryotic release factors
- TCP-seq
References
- ↑ Hellen, Christopher U. T.; Sarnow, Peter (2001-07-01). "Internal ribosome entry sites in eukaryotic mRNA molecules". Genes & Development. 15 (13): 1593–1612. doi:10.1101/gad.891101. ISSN 0890-9369. PMID 11445534.
- ↑ Malys N, McCarthy JEG (2011). "Translation initiation: variations in the mechanism can be anticipated". Cellular and Molecular Life Sciences. 68 (6): 991–1003. doi:10.1007/s00018-010-0588-z. PMID 21076851.
- ↑ Wells, SE; et al. (1998). "Circularization of mRNA by Eukaryotic Translation Initiation Factors". Molecular Cell. 2: 135–140. doi:10.1016/S1097-2765(00)80122-7. PMID 9702200.
- ↑ Cenik, Can; Cenik, Elif Sarinay; Byeon, Gun W; Candille, Sophie P.; Spacek, Damek; Araya, Carlos L; Tang, Hua; Ricci, Emiliano; Snyder, Michael P. (Nov 2015). "Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans.". Genome Research. 25: 1610–21. doi:10.1101/gr.193342.115. PMID 26297486.
- ↑ López-Lastra, M; Rivas, A; Barría, MI (2005). "Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation". Biological research. 38 (2–3): 121–46. doi:10.4067/s0716-97602005000200003. PMID 16238092.
- ↑ Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell. 2007 Sep;99(9):475-87.
- ↑ Schueren F and Thoms S (2016). "Functional Translational Readthrough: A Systems Biology Perspective". PLOS Genetics. 12 (e1006196): 12. doi:10.1371/journal.pgen.100619. PMC 4973966
 . PMID 27490485.
. PMID 27490485.