Post-transcriptional modification

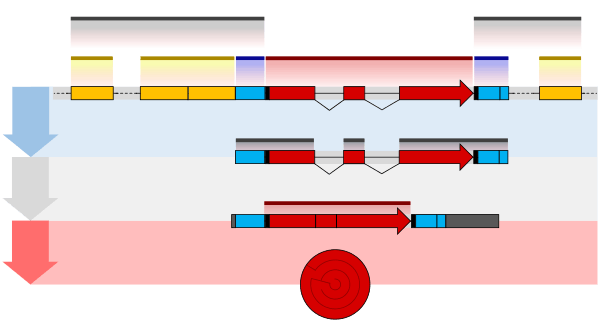
Post-transcriptional modification or Co-transcriptional modification is the process in eukaryotic cells where primary transcript RNA is converted into mature RNA. A notable example is the conversion of precursor messenger RNA into mature messenger RNA (mRNA) that occurs prior to protein translation. The process includes three major steps: addition of a 5' cap, addition of a 3' poly-adenylation tail, and splicing. This process is vital for the correct translation of the genomes of eukaryotes because the initial precursor mRNA produced during transcription contains both exons (coding or important sequences involved in translation), and introns (non-coding sequences).[1]
mRNA processing
|
|
The pre-mRNA molecule undergoes three main modifications. These modifications are 5' capping, 3' polyadenylation, and RNA splicing, which occur in the cell nucleus before the RNA is translated.[2]
5' Processing
Capping
Capping of the pre-mRNA involves the addition of 7-methylguanosine (m7G) to the 5' end. To achieve this, the terminal 5' phosphate requires removal, which is done with the aid of a phosphatase enzyme. The enzyme guanosyl transferase then catalyses the reaction, which produces the diphosphate 5' end. The diphosphate 5' end then attacks the alpha phosphorus atom of a GTP molecule in order to add the guanine residue in a 5'5' triphosphate link. The enzyme (guanine-N7-)-methyltransferase ("cap MTase") transfers a methyl group from S-adenosyl methionine to the guanine ring.[3] This type of cap, with just the (m7G) in position is called a cap 0 structure. The ribose of the adjacent nucleotide may also be methylated to give a cap 1. Methylation of nucleotides downstream of the RNA molecule produce cap 2, cap 3 structures and so on. In these cases the methyl groups are added to the 2' OH groups of the ribose sugar. The cap protects the 5' end of the primary RNA transcript from attack by ribonucleases that have specificity to the 3'5' phosphodiester bonds.[4]
3' Processing
Cleavage and polyadenylation
The pre-mRNA processing at the 3' end of the RNA molecule involves cleavage of its 3' end and then the addition of about 250 adenine residues to form a poly(A) tail. The cleavage and adenylation reactions occur if a polyadenylation signal sequence (5'- AAUAAA-3') is located near the 3' end of the pre-mRNA molecule, which is followed by another sequence, which is usually (5'-CA-3') and is the site of cleavage. A GU-rich sequence is also usually present further downstream on the pre-mRNA molecule. After the synthesis of the sequence elements, two multisubunit proteins called cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CStF) are transferred from RNA Polymerase II to the RNA molecule. The two factors bind to the sequence elements. A protein complex forms that contains additional cleavage factors and the enzyme Polyadenylate Polymerase (PAP). This complex cleaves the RNA between the polyadenylation sequence and the GU-rich sequence at the cleavage site marked by the (5'-CA-3') sequences. Poly(A) polymerase then adds about 200 adenine units to the new 3' end of the RNA molecule using ATP as a precursor. As the poly(A) tail is synthesised, it binds multiple copies of poly(A) binding protein, which protects the 3'end from ribonuclease digestion.[4]
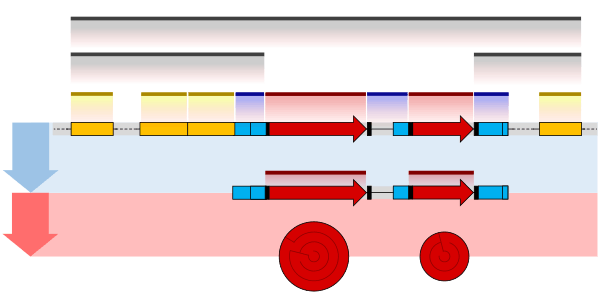
Splicing
RNA splicing is the process by which introns, regions of RNA that do not code for protein, are removed from the pre-mRNA and the remaining exons connected to re-form a single continuous molecule. Exons are sections of mRNA which become "expressed" or translated into a protein, they are the coding portions of a mRNA molecule.[5] Although most RNA splicing occurs after the complete synthesis and end-capping of the pre-mRNA, transcripts with many exons can be spliced co-transcriptionally.[6] The splicing reaction is catalyzed by a large protein complex called the spliceosome assembled from proteins and small nuclear RNA molecules that recognize splice sites in the pre-mRNA sequence. Many pre-mRNAs, including those encoding antibodies, can be spliced in multiple ways to produce different mature mRNAs that encode different protein sequences. This process is known as alternative splicing, and allows production of a large variety of proteins from a limited amount of DNA.
Histone mRNA processing
Histones H2A, H2B, H3 and H4 form the core of a nucleosome and thus are called core histones. Processing of core histones is done differently because typical histone mRNA lacks several features of other eukaryotic mRNAs, such as poly(A) tail and introns. Thus such mRNAs do not undergo splicing and their 3'-prime processing is done independent of most cleavage and polyadenylation factors. Core histone mRNAs have a special stem-loop structure at 3-prime end that is recognized by a stem–loop binding protein and a downstream sequence, called histone downstream element (HDE) that recruits U7 snRNA. Cleavage and polyadenylation specificity factor 73 cuts mRNA between stem-loop and HDE[7]
Histone variants, such as H2A.Z or H3.3, however, have introns and are processed as normal mRNAs including splicing and polyadenylation.[7]
Post-transcriptional modification resources
| Name | Description | Type | Link | References |
|---|---|---|---|---|
| RMBase | RMBase is designed for decoding the landscape of RNA modifications identified from high-throughput sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, miCLIP, m6A-CLIP, RiboMeth-seq). It demonstrated thousands of RNA modifications located within mRNAs, regulatory ncRNAs (e.g. lncRNAs, miRNAs), miRNA target sites and disease-related SNPs. | database | website | [8] |
| MODOMICS | MODOMICS is a database of RNA modifications that provides comprehensive information concerning the chemical structures of modified ribonucleosides, their biosynthetic pathways, RNA-modifying enzymes and location of modified residues in RNA sequences. | database | website | [9] |
| RNAMDB | RNAMDB has served as a focal point for information pertaining to naturally occurring RNA modifications | database | website | [10] |
| . | ||||
Citations
- ↑ Berg, Tymoczko & Stryer 2007, p. 836
- ↑ Berg, Tymoczko & Stryer 2007, p. 841
- ↑ Yamada-Okabe, Toshiko; Mio, Toshiyuki; Matsui, Mitsuaki; Arisawa, Mikio; Yamada-Okabe, Hisafumi (November 1999). "The Candida albicans gene for mRNA 5'-cap methyltransferase: identification of additional residues essential for catalysis". Microbiology. 145 (11): 3023–3033. doi:10.1099/00221287-145-11-3023. ISSN 1350-0872. PMID 10589710. Retrieved January 7, 2011.
- 1 2 Hames & Hooper 2006, p. 221
- ↑ Biology. Mgraw hill education. pp. 241–242. ISBN 978-981-4581-85-1.
- ↑ Lodish HF, Berk A, Kaiser C, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira PT (2007). "Chapter 8: Post-transcriptional Gene Control". Molecular Cell .Biology. San Francisco: WH Freeman. ISBN 0-7167-7601-4.
- 1 2 William F. Marzluff, Eric J. Wagner & Robert J. Duronio (November 2008). "Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail". Nature Reviews Genetics. 9 (11): 843–854. doi:10.1038/nrg2438. PMC 2715827
 . PMID 18927579.
. PMID 18927579. - ↑ Sun, WJ; Li, JH; Liu, S; Wu, J; Zhou, H; Qu, LH; Yang, JH (11 October 2015). "RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data.". Nucleic Acids Research: gkv1036. doi:10.1093/nar/gkv1036. PMID 26464443.
- ↑ Machnicka, MA; Milanowska, K; Osman Oglou, O; Purta, E; Kurkowska, M; Olchowik, A; Januszewski, W; Kalinowski, S; Dunin-Horkawicz, S; Rother, KM; Helm, M; Bujnicki, JM; Grosjean, H (December 2012). "MODOMICS: a database of RNA modification pathways--2013 update.". Nucleic Acids Research. 41 (Database issue): D262–7. doi:10.1093/nar/gks1007. PMID 23118484.
- ↑ Cantara, WA; Crain, PF; Rozenski, J; McCloskey, JA; Harris, KA; Zhang, X; Vendeix, FA; Fabris, D; Agris, PF (December 2010). "The RNA Modification Database, RNAMDB: 2011 update.". Nucleic Acids Research. 39 (Database issue): D195–201. doi:10.1093/nar/gkq1028. PMC 3013656
 . PMID 21071406.
. PMID 21071406.
External links
RMBase RNA Modification Base is designed for decoding the landscape of RNA modifications identified from high-throughput sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, miCLIP, m6A-CLIP, RiboMeth-seq).
- Post-Transcriptional RNA Modification at the US National Library of Medicine Medical Subject Headings (MeSH)
See also
References
- Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2007), Biochemistry (6 ed.), New York: WH Freeman & Co., ISBN 0-7167-6766-X
- Hames, David; Hooper, Nigel (2006), Instant Notes Biochemistry (3 ed.), Leeds: Taylor and Francis, ISBN 0-415-36778-6