Protein O-GlcNAc transferase
| O-GlcNAc transferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 2.4.1.255 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
O-GlcNAc transferase (EC 2.4.1.255, O-GlcNAc transferase, OGTase, O-linked N-acetylglucosaminyltransferase, uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase, protein O-linked beta-N-acetylglucosamine transferase) is an enzyme with systematic name UDP-N-acetyl-D-glucosamine:protein-O-beta-N-acetyl-D-glucosaminyl transferase.[2][3][4][5][6][7] In humans the enzyme is encoded by the OGT gene.[6][8]
Function
O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) catalyzes the addition of a single N-acetylglucosamine in O-glycosidic linkage to serine or threonine residues of intracellular proteins. Since both phosphorylation and O-GlcNAcylation compete for similar serine or threonine residues, the two processes may compete for sites, or they may alter the substrate specificity of nearby sites by steric or electrostatic effects. Two transcript variants encoding cytoplasmic and mitochondrial isoforms have been found for this gene.[8] OGT glycosylates many proteins including: Histone H2B,[9] AKT1,[10] PFKL,[11] KMT2E/MLL5,[11] MAPT/TAU,[12] Host cell factor C1,[13] and SIN3A.[14]
O-GlcNAc transferase is a part of a host of biological functions within the human body. OGT is involved in the resistance of insulin in muscle cells and adipocytes by inhibiting the Threonine 308 phosphorylation of AKT1, increasing the rate of IRS1 phosphorylation (at Serine 307 and Serine 632/635), reducing insulin signaling, and glycosylating components of insulin signals.[15] Additionally, O-GlcNAc transferase catalyzes intracellular glycosylation of serine and threonine residues with the addition of N-acetylglucosamine. Studies show that OGT alleles are vital for embryogenesis, and that OGT is necessary for intracellular glycosylation and embryonic stem cell vitality.[16] O-GlcNAc transferase also catalyzes the posttranslational modification that modifies transcription factors and RNA polymerase II, however the specific function of this modification is mostly unknown.[17]
OGT cleaves Host Cell Factor C1, at one of the 6 repeat sequences. The TPR repeat domain of OGT binds to the carboxyl terminal portion of an HCF1 proteolytic repeat so that the cleavage region is in the glycosyltransferase active site above uridine-diphosphate-GlcNAc [7] The large proportion of OGT complexed with HCF1 is necessary for HCF1 cleavage, and HCFC1 is required for OGT stabilization in the nucleus. HCF1 regulates OGT stability using a post-transcriptional mechanism, however the mechanism of the interaction with HCFC1 is still unknown.[18]
Structure
The human OGT gene has 1046 amino acid residues, and is a heterotrimer consisting of two 110 kDa subunits and one 78 kDa subunit.[5] The 110 kDa subunit contains 13 tetratricopeptide (TPR) repeats; the 13th repeat is truncated. These subunits are dimerized by TPR repeats 6 and 7. OGT is highly expressed in the pancreas and also expressed in the heart, brain, skeletal muscle, and the placenta. There have been trace amounts found in the lung and the liver.[6] The binding sites have been determined for the 110 kDa subunit. It has 3 binding sites at amino acid residues 849, 852, and 935. The probable active site is at residue 508.[11]
The crystal structure of O-GlcNAc transferase has not been well studied, but the structure of a binary complex with UDP and a ternary complex with UDP and a peptide substrate has been researched.[7] The OGT-UDP complex contains three domains in its catalytic region: the amino (N)-terminal domain, the carboxy (C)-terminal domain, and the intervening domain (Int-D). The catalytic region is linked to TPR repeats by a translational helix (H3), which loops from the C-cat domain to the N-Cat domain along the upper surface of the catalytic region.[7] The OGT-UDP-peptide complex has a larger space between the TPR domain and the catalytic region than the OGT-UDP complex. The CKII peptide, which contains three serine residues and a threonine residue, binds in this space. This structure supports an ordered sequential bi-bi mechanism that matches the fact that “at saturating peptide concentrations, a competitive inhibition pattern was obtained for UDP with respect to UDP-GlcNAc.”[7]
Mechanism of catalysis
The molecular mechanism of O-linked N-acetylglucosamine transferase has not been extensively studied either, since there is not a confirmed crystal structure of the enzyme. A proposed mechanism by Lazarus et al. is supported by product inhibition patterns of UDP at saturating peptide conditions. This mechanism proceeds with starting materials Uridine diphosphate N-acetylglucosamine, and a peptide chain with a reactive serine or threonine hydroxyl group. The proposed reaction is an ordered sequential bi-bi mechanism.[7]

The chemical reaction can be written as:
(1) UDP-N-acetyl-D-glucosamine + [protein]-L-serine → UDP + [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine
(2) UDP-N-acetyl-D-glucosamine + [protein]-L-threonine → UDP + [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-threonine
First, the hydroxyl group of serine is deprotonated by Histidine 498, a catalytic base in this proposed reaction. Lysine 842 is also present to stabilize the UDP moiety. The oxygen ion then attacks the sugar-phosphate bond between the glucosamine and UDP. This results in the splitting of UDP-N-acetylglucosamine into N-acetylglucosamine – Peptide and UDP. Proton transfers take place at the phosphate and Histidine 498. This mechanism is spurred by OGT gene containing O-linked N-acetylglucosamine transferase. Aside from proton transfers the reaction proceeds in one step, as shown in Figure 2.[7] Figure 2 uses a lone serine residue as a representative of the peptide with a reactive hydroxyl group. Threonine could have also been used in the mechanism.
Regulation
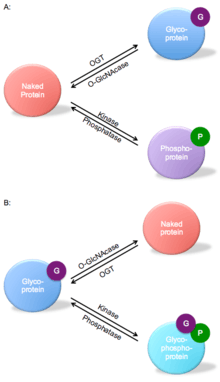
O-GlcNAc transferase is part of a dynamic competition for a serine or threonine hydroxyl functional group in a peptide unit. Figure 3 shows an example of both reciprocal same-site occupancy and adjacent-site occupancy. For the same-site occupancy, OGT competes with kinase to catalyze the glycosylation of the protein instead of phosphorylation. The adjacent-site occupancy example shows the naked protein catalyzed by OGT converted to a glycoprotein, which can increase the turnover of proteins such as the tumor repressor p53.[19]
The post-translational modification of proteins by O-GlcNAc is spurred by glucose flux through the hexosamine biosynthetic pathway. OGT catalyzes attachment of the O-GlcNAc group to serine and threonine, while O-GlcNAcase spurs sugar removal.[20][21]
This regulation is important for multiple cellular processes including transcription, signal transduction, and proteasomal degradation. Also, there is competitive regulation between OGT and kinase for the protein to attach to a phosphate group or O-GlcNAc, which can alter the function of proteins in the body through downstream effects.[11][20] OGT inhibits the activity of 6-phosophofructosekinase PFKL by mediating the glycosylation process. This then acts as a part of glycolysis regulation. O-GlcNAc has been defined as a negative transcription regulator in response to steroid hormone signaling.[15]
Studies show that O-GlcNAc transferase interacts directly with the Ten eleven translocation 2 (TET2) enzyme, which converts 5-methylcytosine to 5-hydroxymethylcytosine and regulates gene transcription.[22] Additionally, increasing levels of OGT for O-GlcNAcylation may have therapeutic effects for Alzheimer's disease patients. Brain glucose metabolism is impaired in Alzheimer's disease, and a study suggests that this leads to hyperphosphorylation of tau and degerenation of tau O-GlcNCAcylation. Replenishing tau O-GlcNacylation in the brain along with protein phosphatase could deter this process and improve brain glucose metabolism.[12]
References
- ↑ Jinek, M; Rehwinkel, J; Lazarus, B; Izaurraide, E; Hanover, J; Conti, E (12 September 2004). "The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha". Nature Structural & Molecular Biology. 11 (10): 1001–1007. doi:10.1038/nsmb833. PMID 15361863.
- ↑ Banerjee, S.; Robbins, P.W.; Samuelson, J. (2009). "Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidium parvum". Glycobiology. 19 (4): 331–336. doi:10.1093/glycob/cwn107. PMID 18948359.
- ↑ Clarke, A.J.; Hurtado-Guerrero, R.; Pathak, S.; Schuttelkopf, A.W.; Borodkin, V.; Shepherd, S.M.; Ibrahim, A.F.; van Aalten, D.M. (2008). "Structural insights into mechanism and specificity of O-GlcNAc transferase". EMBO J. 27 (20): 2780–2788. doi:10.1038/emboj.2008.186. PMID 18818698.
- ↑ Rao, F.V.; Dorfmueller, H.C.; Villa, F.; Allwood, M.; Eggleston, I.M.; van Aalten, D.M. (2006). "Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis". EMBO J. 25 (7): 1569–1578. doi:10.1038/sj.emboj.7601026. PMID 16541109.
- 1 2 Haltiwanger, R.S.; Blomberg, M.A.; Hart, G.W. (1992). "Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase". J. Biol. Chem. 267 (13): 9005–9013. PMID 1533623.
- 1 2 3 Lubas, W.A.; Frank, D.W.; Krause, M.; Hanover, J.A. (1997). "O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats". J. Biol. Chem. 272: 9316–9324. doi:10.1074/jbc.272.14.9316. PMID 9083068.
- 1 2 3 4 5 6 7 8 Lazarus, M.B.; Nam, Y.; Jiang, J.; Sliz, P.; Walker, S. (2011). "Structure of human O-GlcNAc transferase and its complex with a peptide substrate". Nature. 469 (7331): 564–567. doi:10.1038/nature09638. PMC 3064491
 . PMID 21240259.
. PMID 21240259. - 1 2 "Entrez Gene: OGT O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)".
- ↑ Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S (December 2011). "GlcNAcylation of histone H2B facilitates its monoubiquitination". Nature. 480 (7378): 557–60. doi:10.1038/nature10656. PMID 22121020.
- ↑ Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW (February 2010). "Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes". J. Biol. Chem. 285 (8): 5204–11. doi:10.1074/jbc.M109.077818. PMC 2820748
 . PMID 20018868.
. PMID 20018868. - 1 2 3 4 "O15294 (OGT1_HUMAN) Reviewed, UniProtKB/Swiss-Prot". UniProt.
- 1 2 Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, Gong CX (July 2009). "Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease". Brain. 132 (Pt 7): 1820–32. doi:10.1093/brain/awp099. PMC 2702834
 . PMID 19451179.
. PMID 19451179. - ↑ Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W (April 2003). "Human Sin3 deacetylase trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1". Genes Dev. 17 (7): 896–911. doi:10.1101/gad.252103. PMC 196026
 . PMID 12670868.
. PMID 12670868. - ↑ Yang X, Zhang F, Kudlow JE (July 2002). "Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression". Cell. 110 (1): 69–80. doi:10.1016/S0092-8674(02)00810-3. PMID 12150998.
- 1 2 Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM (February 2008). "Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance". Nature. 451 (7181): 964–9. doi:10.1038/nature06668. PMID 18288188.
- ↑ Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD (May 2000). "The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny". Proc. Natl. Acad. Sci. U.S.A. 97 (11): 5735–9. doi:10.1073/pnas.100471497. PMC 18502
 . PMID 10801981.
. PMID 10801981. - ↑ Chen Q, Chen Y, Bian C, Fujiki R, Yu X (January 2013). "TET2 promotes histone O-GlcNAcylation during gene transcription". Nature. 493 (7433): 561–4. doi:10.1038/nature11742. PMC 3684361
 . PMID 23222540.
. PMID 23222540. - ↑ Daou S, Mashtalir N, Hammond-Martel I, Pak H, Yu H, Sui G, Vogel JL, Kristie TM, Affar el B (February 2011). "Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway". Proc. Natl. Acad. Sci. U.S.A. 108 (7): 2747–52. doi:10.1073/pnas.1013822108. PMC 3041071
 . PMID 21285374.
. PMID 21285374. - 1 2 Hart GW, Housley MP, Slawson C (April 2007). "Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins". Nature. 446 (7139): 1017–22. doi:10.1038/nature05815. PMID 17460662.
- 1 2 Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE (June 2001). "O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability". Proc. Natl. Acad. Sci. U.S.A. 98 (12): 6611–6. doi:10.1073/pnas.111099998. PMC 34401
 . PMID 11371615.
. PMID 11371615. - ↑ Love DC, Hanover JA (November 2005). "The hexosamine signaling pathway: deciphering the "O-GlcNAc code"". Sci. STKE. 2005 (312): re13. doi:10.1126/stke.3122005re13. PMID 16317114.
- ↑ Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (May 2009). "Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1". Science. 324 (5929): 930–5. doi:10.1126/science.1170116. PMC 2715015
 . PMID 19372391.
. PMID 19372391.
External links
- Protein O-GlcNAc transferase at the US National Library of Medicine Medical Subject Headings (MeSH)