Methylenetetrahydrofolate reductase
| View/Edit Human | View/Edit Mouse |
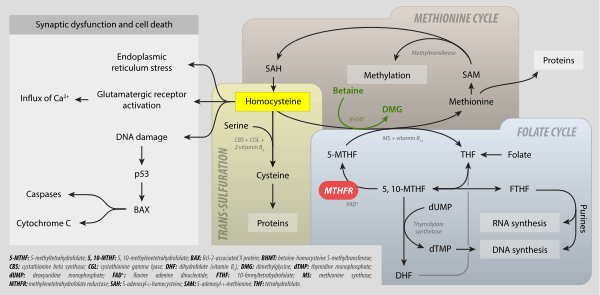
Methylene tetrahydrofolate reductase (MTHFR) is the rate-limiting enzyme in the methyl cycle, and it is encoded by the MTHFR gene.[3] Methylenetetrahydrofolate reductase catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a cosubstrate for homocysteine remethylation to methionine. Natural variation in this gene is common in healthy people. Although some variants have been reported to influence susceptibility to occlusive vascular disease, neural tube defects, Alzheimer's disease and other forms of dementia, colon cancer, and acute leukemia, findings from small early studies have not been reproduced. Some mutations in this gene are associated with methylene tetrahydrofolate reductase deficiency.[4][5][6]
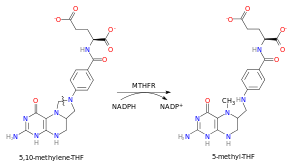
Biochemistry
| methylene tetrahydrofolate reductase [NAD(P)H] | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Schematic diagram of the reductive carbon-nitrogen bond cleavage (represented by wavy line) catalyzed by methylenetetrahydrofolate reductase. | |||||||||
| Identifiers | |||||||||
| EC number | 1.5.1.20 | ||||||||
| CAS number | 9028-69-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In the rate-limiting step of the methyl cycle, MTHFR irreversibly reduces 5,10-methylenetetrahydrofolate (substrate) to 5-methyltetrahydrofolate (product).
- 5,10-methylene tetrahydrofolate is used to convert dUMP to dTMP for de novo thymidine synthesis.
- 5-Methyltetrahydrofolate is used to convert homocysteine (a potentially toxic amino acid) to methionine by the enzyme methionine synthase. (Note that homocysteine can also be converted to methionine by the folate-independent enzyme betaine-homocysteine methyltransferase (BHMT))
MTHFR contains a bound flavin cofactor and uses NAD(P)H as the reducing agent.
Structure
Mammalian MTHFR is composed of an N-terminal catalytic domain and a C-terminal regulatory domain. MTHFR has at least two promoters and two isoforms (70 kDa and 77 kDa).[7]
Regulation
MTHFR activity may be inhibited by binding of dihydrofolate (DHF)[8] and S-adenosylmethionine (SAM, or AdoMet).[9] MTHFR can also be phosphorylated – this decreases its activity by ~20% and allows it to be more easily inhibited by SAM.[10]
Genetics
The enzyme is coded by the gene with the symbol MTHFR on chromosome 1 location p36.3 in humans.[11] There are DNA sequence variants (genetic polymorphisms) associated with this gene. In 2000 a report brought the number of polymorphisms up to 24.[12] Two of the most investigated are C677T (rs1801133) and A1298C (rs1801131) single nucleotide polymorphisms (SNP).
C677T SNP (Ala222Val)
The MTHFR nucleotide at position 665 in the gene has two possibilities: C (cytosine) or T (thymine). C at position 665 (leading to an alanine at amino acid 222) is the normal allele. The 665T allele (leading to a valine substitution at amino acid 222) encodes a thermolabile enzyme with reduced activity.
Individuals with two copies of 665C (665CC) have the most common genotype. 665TT individuals (homozygous) have lower MTHFR activity than CC or CT (heterozygous) individuals. About ten percent of the North American population are T-homozygous for this polymorphism. There is ethnic variability in the frequency of the T allele – frequency in Mediterranean/Hispanics is greater than the frequency in Caucasians which, in turn, is greater than in Africans/African-Americans.[13]
The degree of enzyme thermolability (assessed as residual activity after heat inactivation) is much greater in 665TT individuals (18-22%) compared with 677CT (56%) and 677CC (66-67%).[14] Individuals of 677TT are predisposed to mild hyperhomocysteinemia (high blood homocysteine levels), because they have less active MTHFR available to produce 5-methyltetrahydrofolate (which is used to decrease homocysteine). Low dietary intake of the vitamin folic acid can also cause mild hyperhomocysteinemia.
Low folate intake affects individuals with the 677TT genotype to a greater extent than those with the 677CC/CT genotypes. 677TT (but not 677CC/CT) individuals with lower plasma folate levels are at risk for elevated plasma homocysteine levels.[15] In studies of human recombinant MTHFR, the protein encoded by 677T loses its FAD cofactor three times faster than the wild-type protein.[16] 5-Methyl-THF slows the rate of FAD release in both the wild-type and mutant enzymes, although it is to a much greater extent in the mutant enzyme.<ref name="YamadaK2001Effects"/ Low folate status with the consequent loss of FAD enhances the thermolability of the enzyme, thus providing an explanation for the normalised homocysteine and DNA methylation levels in folate-replete 677TT individuals.
This polymorphism and mild hyperhomocysteinemia are associated with neural tube defects in offspring, increased risk for [http://www.rbej.com/content/2/1/65 complications of pregnancy other complications of pregnancy, arterial and venous thrombosis, and cardiovascular disease.[17] 677TT individuals are at an increased risk for acute lymphoblastic leukemia[18] and colon cancer.[19]
Mutations in the MTHFR gene could be one of the factors leading to increased risk of developing schizophrenia.[20] Schizophrenic patients having the risk allele (T\T) show more deficiencies in executive function tasks.[21]
The C677T genotype is associated with increased risk of recurrent pregnancy loss (RPL) in non Caucasians.[22]
There is also a tentative link between MTHFR mutations and dementia. One study of an elderly Japanese population[23] found correlations between the MTHFR 677CT mutation, an Apo E polymorphism, and certain types of senile dementia. Other research has found that individuals with folate-related mutations can still have a functional deficiency even when blood levels of folate are within the normal range,[24] and recommended supplementation of methyltetrahydrofolate to potentially prevent and treat dementia (along with depression). A 2011 study[25] from China also found that the C677T SNP was associated with Alzheimer's disease in Asian populations (though not in Caucasians).
A1305C SNP (Glu429Ala)
At nucleotide 1305 of the MTHFR, there are two possibilities: A or C. 1305A (leading to a Glu at amino acid 429) is the most common while 1305C (leading to an Ala substitution at amino acid 429) is less common. 1305AA is the "normal" homozygous, 1305AC the heterozygous, and 1298CC the homozygous for the "variant". In studies of human recombinant MTHFR, the protein encoded by 1305C cannot be distinguished from 1305A in terms of activity, thermolability, FAD release, or the protective effect of 5-methyl-THF.[16] The C mutation does not appear to affect the MTHFR protein. It does not result in thermolabile MTHFR and does not appear to affect homocysteine levels. It does, however, affect the conversion of MTHF to BH4 (tetrahydrobiopterin), an important cofactor in the production of neurotransmitters, synthesis of nitric oxide, and detoxification of ammonia.
There has been some commentary on a 'reverse reaction' in which tetrahydrobiopterin (BH4) is produced when 5-methyltetrahydrofolate is converted back into methylenetetrahydrofolate. This however is not universally agreed upon. That reaction is thought to require 5-MTHF and SAMe. An alternative opinion is that 5-MTHF processes peroxynitrite, thereby preserving existing BH4, and that no such 'reverse reaction' occurs.
Detection of MTHFR polymorphisms
A triplex tetra-primer ARMS-PCR method was developed for the simultaneous detection of C677T and A1298C polymorphisms with the A66G MTRR polymorphism in a single PCR reaction.[26]
Severe MTHFR deficiency
Severe MTHFR deficiency is rare (about 50 cases worldwide) and caused by mutations resulting in 0-20% residual enzyme activity.[12] Patients exhibit developmental delay, motor and gait dysfunction, seizures, and neurological impairment and have extremely high levels of homocysteine in their plasma and urine as well as low to normal plasma methionine levels.
As a drug target
Inhibitors of MTHFR and antisense knockdown of the expression of the enzyme have been proposed as treatments for cancer.[27] The active form of folate, L-methylfolate, may be appropriate to target for conditions affected by MTHFR polymorphisms.[28]
Reaction and metabolism
The overall reaction catalyzed by MTHFR is illustrated on the right. The reaction uses an NAD(P)H hydride donor and an FAD cofactor. The E. coli enzyme has a strong preference for the NADH donor, whereas the mammalian enzyme is specific to NADPH.
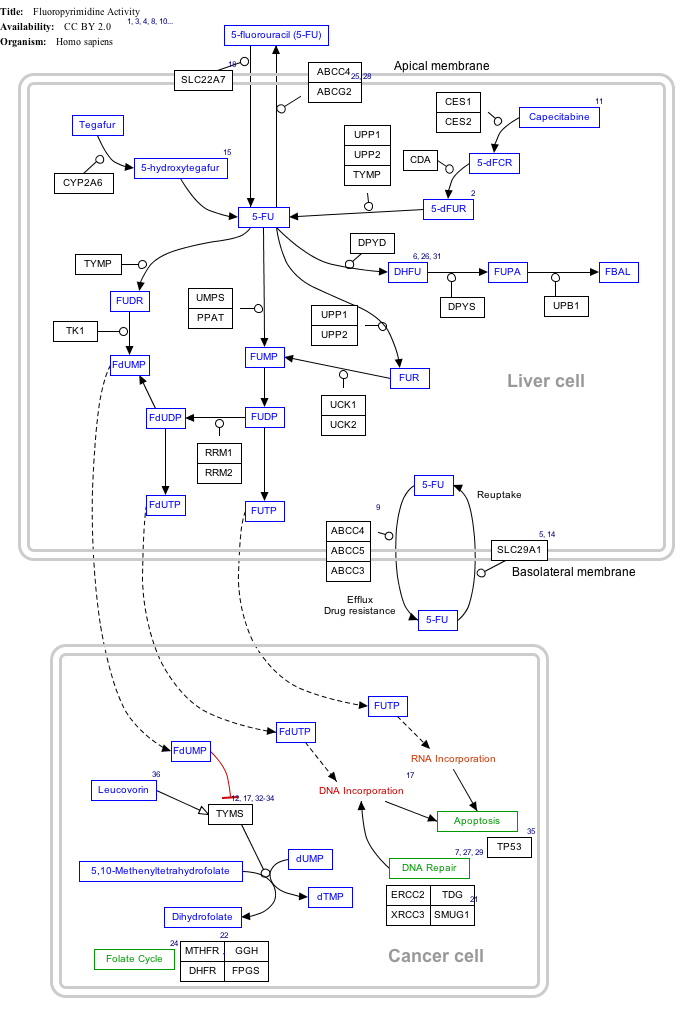
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
Fluorouracil (5-FU) Activity edit
- ↑ The interactive pathway map can be edited at WikiPathways: "FluoropyrimidineActivity_WP1601".
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R (Jun 1994). "Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification". Nature Genetics. 7 (2): 195–200. doi:10.1038/ng0694-195. PMID 7920641.
- ↑ "Entrez Gene: MTHFR methylene tetrahydrofolate reductase (NAD(P)H)".
- ↑ Födinger M, Hörl WH, Sunder-Plassmann G (2000). "Molecular biology of 5,10-methylenetetrahydrofolate reductase". Journal of Nephrology. 13 (1): 20–33. PMID 10720211.
- ↑ Trimmer EE (2013). "Methylenetetrahydrofolate reductase: biochemical characterization and medical significance". Current Pharmaceutical Design. 19 (14): 2574–93. doi:10.2174/1381612811319140008. PMID 23116396.
- ↑ Tran P, Leclerc D, Chan M, Pai A, Hiou-Tim F, Wu Q, Goyette P, Artigas C, Milos R, Rozen R (Sep 2002). "Multiple transcription start sites and alternative splicing in the methylenetetrahydrofolate reductase gene result in two enzyme isoforms". Mammalian Genome. 13 (9): 483–92. doi:10.1007/s00335-002-2167-6. PMID 12370778.
- ↑ Matthews RG, Daubner SC (1982). "Modulation of methylenetetrahydrofolate reductase activity by S-adenosylmethionine and by dihydrofolate and its polyglutamate analogues". Advances in Enzyme Regulation. 20: 123–31. doi:10.1016/0065-2571(82)90012-7. PMID 7051769.
- ↑ Jencks DA, Mathews RG (Feb 1987). "Allosteric inhibition of methylenetetrahydrofolate reductase by adenosylmethionine. Effects of adenosylmethionine and NADPH on the equilibrium between active and inactive forms of the enzyme and on the kinetics of approach to equilibrium". The Journal of Biological Chemistry. 262 (6): 2485–93. PMID 3818603.
- ↑ Yamada K, Strahler JR, Andrews PC, Matthews RG (Jul 2005). "Regulation of human methylenetetrahydrofolate reductase by phosphorylation". Proceedings of the National Academy of Sciences of the United States of America. 102 (30): 10454–9. doi:10.1073/pnas.0504786102. PMC 1180802
 . PMID 16024724.
. PMID 16024724. - ↑ Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R (Aug 1994). "Human methylenetetrahydrofolate reductase: isolation of cDNA mapping and mutation identification". Nature Genetics. 7 (4): 551. doi:10.1038/ng0894-551a. PMID 7951330.
- 1 2 Sibani S, Christensen B, O'Ferrall E, Saadi I, Hiou-Tim F, Rosenblatt DS, Rozen R (2000). "Characterization of six novel mutations in the methylenetetrahydrofolate reductase (MTHFR) gene in patients with homocystinuria". Human Mutation. 15 (3): 280–7. doi:10.1002/(SICI)1098-1004(200003)15:3<280::AID-HUMU9>3.0.CO;2-I. PMID 10679944.
- ↑ Schneider JA, Rees DC, Liu YT, Clegg JB (May 1998). "Worldwide distribution of a common methylenetetrahydrofolate reductase mutation". American Journal of Human Genetics. 62 (5): 1258–60. doi:10.1086/301836. PMC 1377093
 . PMID 9545406.
. PMID 9545406. - ↑ Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP (May 1995). "A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase". Nature Genetics. 10 (1): 111–3. doi:10.1038/ng0595-111. PMID 7647779.
- ↑ Reilly R, McNulty H, Pentieva K, Strain JJ, Ward M (Feb 2014). "MTHFR 677TT genotype and disease risk: is there a modulating role for B-vitamins?". The Proceedings of the Nutrition Society. 73 (1): 47–56. doi:10.1017/S0029665113003613. PMID 24131523.
- 1 2 Yamada K, Chen Z, Rozen R, Matthews RG (Dec 2001). "Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase". Proceedings of the National Academy of Sciences of the United States of America. 98 (26): 14853–8. doi:10.1073/pnas.261469998. PMC 64948
 . PMID 11742092.
. PMID 11742092. - ↑ Schwahn B, Rozen R (2001). "Polymorphisms in the methylenetetrahydrofolate reductase gene: clinical consequences". American Journal of Pharmacogenomics. 1 (3): 189–201. doi:10.2165/00129785-200101030-00004. PMID 12083967.
- ↑ Ojha RP, Gurney JG (Jan 2014). "Methylenetetrahydrofolate reductase C677T and overall survival in pediatric acute lymphoblastic leukemia: a systematic review". Leukemia & Lymphoma. 55 (1): 67–73. doi:10.3109/10428194.2013.792336. PMID 23550988.
- ↑ Bailey LB (Nov 2003). "Folate, methyl-related nutrients, alcohol, and the MTHFR 677C-->T polymorphism affect cancer risk: intake recommendations". The Journal of Nutrition. 133 (11 Suppl 1): 3748S–3753S. PMID 14608109.
- ↑ "Meta-Analysis of All Published Schizophrenia-Association Studies (Case-Control Only) for rs1801133 (C677T) polymorphism, MTHFR gene". Schizophrenia Research Forum. Retrieved 2007-03-11.
- ↑ Roffman JL, Weiss AP, Deckersbach T, Freudenreich O, Henderson DC, Purcell S, Wong DH, Halsted CH, Goff DC (May 2007). "Effects of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism on executive function in schizophrenia". Schizophrenia Research. 92 (1-3): 181–8. doi:10.1016/j.schres.2007.01.003. PMID 17344026.
- ↑ Wu X, Zhao L, Zhu H, He D, Tang W, Luo Y (Jul 2012). "Association between the MTHFR C677T polymorphism and recurrent pregnancy loss: a meta-analysis". Genetic Testing and Molecular Biomarkers. 16 (7): 806–11. doi:10.1089/gtmb.2011.0318. PMID 22313097.
- ↑ Nishiyama M, Kato Y, Hashimoto M, Yukawa S, Omori K (May 2000). "Apolipoprotein E, methylenetetrahydrofolate reductase (MTHFR) mutation and the risk of senile dementia--an epidemiological study using the polymerase chain reaction (PCR) method". Journal of Epidemiology / Japan Epidemiological Association. 10 (3): 163–72. doi:10.2188/jea.10.163. PMID 10860300.
- ↑ Mischoulon D, Raab MF (2007). "The role of folate in depression and dementia". The Journal of Clinical Psychiatry. 68 Suppl 10: 28–33. PMID 17900207.
- ↑ Hua Y, Zhao H, Kong Y, Ye M (Aug 2011). "Association between the MTHFR gene and Alzheimer's disease: a meta-analysis". The International Journal of Neuroscience. 121 (8): 462–71. doi:10.3109/00207454.2011.578778. PMID 21663380.
- ↑ Lajin B, Alachkar A, Sakur AA (Feb 2012). "Triplex tetra-primer ARMS-PCR method for the simultaneous detection of MTHFR c.677C>T and c.1298A>C, and MTRR c.66A>G polymorphisms of the folate-homocysteine metabolic pathway". Molecular and Cellular Probes. 26 (1): 16–20. doi:10.1016/j.mcp.2011.10.005. PMID 22074746.
- ↑ Stankova J, Lawrance AK, Rozen R (2008). "Methylenetetrahydrofolate reductase (MTHFR): a novel target for cancer therapy". Current Pharmaceutical Design. 14 (11): 1143–50. doi:10.2174/138161208784246171. PMID 18473861.
- ↑ Papakostas GI, Shelton RC, Zajecka JM, Bottiglieri T, Roffman J, Cassiello C, Stahl SM, Fava M (Aug 2014). "Effect of adjunctive L-methylfolate 15 mg among inadequate responders to SSRIs in depressed patients who were stratified by biomarker levels and genotype: results from a randomized clinical trial". The Journal of Clinical Psychiatry. 75 (8): 855–63. doi:10.4088/JCP.13m08947. PMID 24813065.
Further reading
- Hickey SE, Curry CJ, Toriello HV (Feb 2013). "ACMG Practice Guideline: lack of evidence for MTHFR polymorphism testing". Genetics in Medicine. 15 (2): 153–6. doi:10.1038/gim.2012.165. PMID 23288205.
- Matthews RG (2003). "Methylenetetrahydrofolate reductase: a common human polymorphism and its biochemical implications". Chemical Record. 2 (1): 4–12. doi:10.1002/tcr.10006. PMID 11933257.
- Schwahn B, Rozen R (2002). "Polymorphisms in the methylenetetrahydrofolate reductase gene: clinical consequences". American Journal of Pharmacogenomics. 1 (3): 189–201. doi:10.2165/00129785-200101030-00004. PMID 12083967.
- Iqbal MP, Frossard PM (Jan 2003). "Methylene tetrahydrofolate reductase gene and coronary artery disease". JPMA. the Journal of the Pakistan Medical Association. 53 (1): 33–6. PMID 12666851.
- Bailey LB (Nov 2003). "Folate, methyl-related nutrients, alcohol, and the MTHFR 677C-->T polymorphism affect cancer risk: intake recommendations". The Journal of Nutrition. 133 (11 Suppl 1): 3748S–3753S. PMID 14608109.
- Wiwanitkit V (Jul 2005). "Roles of methylenetetrahydrofolate reductase C677T polymorphism in repeated pregnancy loss". Clinical and Applied Thrombosis/Hemostasis. 11 (3): 343–5. doi:10.1177/107602960501100315. PMID 16015422.
- Muntjewerff JW, Kahn RS, Blom HJ, den Heijer M (Feb 2006). "Homocysteine, methylenetetrahydrofolate reductase and risk of schizophrenia: a meta-analysis". Molecular Psychiatry. 11 (2): 143–9. doi:10.1038/sj.mp.4001746. PMID 16172608.
- Lewis SJ, Lawlor DA, Davey Smith G, Araya R, Timpson N, Day IN, Ebrahim S (Apr 2006). "The thermolabile variant of MTHFR is associated with depression in the British Women's Heart and Health Study and a meta-analysis". Molecular Psychiatry. 11 (4): 352–60. doi:10.1038/sj.mp.4001790. PMID 16402130.
- Pereira TV, Rudnicki M, Pereira AC, Pombo-de-Oliveira MS, Franco RF (Oct 2006). "5,10-Methylenetetrahydrofolate reductase polymorphisms and acute lymphoblastic leukemia risk: a meta-analysis". Cancer Epidemiology, Biomarkers & Prevention. 15 (10): 1956–63. doi:10.1158/1055-9965.EPI-06-0334. PMID 17035405.
- Leclerc D, Rozen R (Mar 2007). "[Molecular genetics of MTHFR: polymorphisms are not all benign]". Médecine Sciences. 23 (3): 297–302. doi:10.1051/medsci/2007233297. PMID 17349292.