Phosphatidylethanolamine
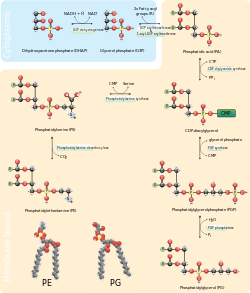
Phosphatidylethanolamines (sometimes abbreviated PE) are a class of phospholipids found in biological membranes.[1] They are synthesized by the addition of CDP-ethanolamine to diglycerides, releasing CMP. S-Adenosyl methionine can subsequently methylate the amine of phosphatidylethanolamines to yield phosphatidylcholines. It can mainly be found in the inner (cytoplasmic) leaflet of the lipid bilayer.[2]
Function

PtdCho - Phosphatidylcholine;
PtdEtn - Phosphatidylethanolamine;
PtdIns -Phosphatidylinositol;
PtdSer - Phosphatidylserine.
In cells
Phosphatidylethanolamines are found in all living cells, composing 25% of all phospholipids. In human physiology, they are found particularly in nervous tissue such as the white matter of brain, nerves, neural tissue, and in spinal cord, where they make up 45% of all phospholipids.[3]
PEs play a role in membrane fusion and in disassembly of the contractile ring during cytokinesis in cell division.[4] Additionally, it is thought that PE regulates membrane curvature. PE acts as an important precursor, substrate, or donor in several biological pathways.[3]
As a polar head group, PE creates a more viscous lipid membrane compared to phosphatidylcholine (PC). For example, the melting temperature of di-oleoyl-PE is -16 °C while the melting temperature of di-oleoyl-PC is -20 °C. If the lipids had two palmitoyl chains, PE would melt at 63°C while PC would melt already at 41 °C.[5] Lower melting temperatures correspond, in a simplistic view, to more fluid membranes.
In humans
In humans, metabolism of PE is thought to be important in the heart. When blood flow to the heart is restricted, the asymmetrical distribution of PE between membrane leaflets is disrupted, and as a result the membrane is disrupted. Additionally, PE plays a role in the secretion of lipoproteins in the liver. This is because vesicles for secretion of VLDLs coming off of the Golgi have a significantly higher PE concentration when compared to other vesicles containing VLDLs.[6] PE has also shown to be able to propagate infectious prions without the assistance of any proteins or nucleic acids, which is a unique characteristic of it.[7] PE is also thought to play a role in blood clotting, as it works with phosphatidylserine to increase the rate of thrombin formation by promoting binding to Factor V and Factor X, two proteins which catalyze the formation of thrombin from prothrombin.[8]
In bacteria
Where phosphatidylcholine is the principal phospholipid in animals, PE is the principal one in bacteria. One of the primary roles for PE in bacterial membranes is to spread out the negative charge caused by anionic membrane phospholipids. In the bacterium E. coli, PE play a role in supporting lactose permease's active transport of lactose into the cell, and may play a role in other transport systems as well. PE plays a role in the assembly of lactose permease and other membrane proteins. It acts as a 'chaperone' to help the membrane proteins correctly fold their tertiary structures so that they can function properly. When PE is not present, the transport proteins have incorrect tertiary structures and do not function correctly.[9]
PE also enables bacterial multidrug transporters to function properly. PE allows the formation of intermediates that are needed for the transporters to properly open and close.[10]
Structure
As a lecithin, PE consists of a combination of glycerol esterified with two fatty acids and phosphoric acid. Whereas the phosphate group is combined with choline in phosphatidylcholine, it is combined with the ethanolamine in PE. The two fatty acids may be the same, or different, and are usually in the 1,2 positions (though they can be in the 1,3 positions).
Synthesis
The phosphatidylserine decarboxylation pathway and the CDP-ethanolamine pathways are used to synthesize PE. Phosphatidylserine decarboxylase (PSD) is the enzyme that is used to decarboxylate phosphatidylserine in the first pathway. The phosphatidylserine decarboxylation pathway is the main source of synthesis for PE in the membranes of the mitochondria. PE produced in the mitochondrial membrane is also transported throughout the cell to other membranes for use. In a process that mirrors phosphatidylcholine synthesis, PE is also made via the CDP-ethanolamine pathway, using ethanolamine as the substrate. Through several steps taking place is both the cytosol and endoplasmic reticulum, the synthesis pathway yields the end product of PE.[11] PE is also found abundantly in soya or egg lecithin and is produced commercially using chromatographic separation.
Regulation
Synthesis of PE through the phosphatidylserine decarboxylation pathway occurs rapidly in the inner mitochondrial membrane. However, phosphatidylserine is made in the endoplasmic reticulum. Because of this, the transport of phosphatidylserine from the endoplasmic reticulum to the mitrochondrial membrane and then to the inner mitochondrial membrane limits the rate of synthesis via this pathway. The mechanism for this transport is currently unknown, but may play a role in regulation of the rate of synthesis in this pathway.
Presence in food, health issues
Phosphatidylethanolamines in food break down to form PE-linked Amadori products as a part of the Maillard reaction.[12] These products accelerate membrane lipid peroxidation, causing oxidative stress to cells that come in contact with them.[13] Oxidative stress is known to cause food deterioration and several diseases. Significant levels of Amadori-PE products have been found in a wide variety of foods such as chocolate, soybean milk, infant formula, and other processed foods. The levels of Amadori-PE products are higher in foods with high lipid and sugar concentrations that have high temperatures in processing.[12] Additional studies have found that Amadori-PE may play a role in vascular disease,[14] act as the mechanism by which diabetes can increase the incidence of cancer,[15] and potentially play a role in other diseases as well. Amadori-PE has a higher plasma concentration in diabetes patients than healthy people, indicating it may play a role in the development of the disease or be a product of the disease.[16]
See also
References
- ↑ Wellner, Niels; Diep, Thi Ai; Janfelt, Christian; Hansen, Harald Severin (2012). "N-acylation of phosphatidylethanolamine and its biological functions in mammals". Biochimica et Biophysica Acta. 1831 (3): 652–62. doi:10.1016/j.bbalip.2012.08.019. PMID 23000428.
- ↑ Mishkind, Michael (2000). "Phosphatidylethanolamine – in a pinch". Trends in Cell Biology. 10 (9): 368. doi:10.1016/S0962-8924(00)01826-2.
- 1 2 Vance, Jean E.; Tasseva, Guergana (2012). "Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells". Biochimica et Biophysica Acta. 1831 (3): 543–54. doi:10.1016/j.bbalip.2012.08.016. PMID 22960354.
- ↑ Emoto, K.; Kobayashi, T; Yamaji, A; Aizawa, H; Yahara, I; Inoue, K; Umeda, M (1996). "Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis". Proceedings of the National Academy of Sciences. 93 (23): 12867–72. Bibcode:1996PNAS...9312867E. doi:10.1073/pnas.93.23.12867. JSTOR 40713. PMC 24012
 . PMID 8917511.
. PMID 8917511. - ↑ See references in Wan et al. Biochemistry 47 2008
- ↑ Vance, J. E. (2008). "Thematic Review Series: Glycerolipids. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: Two metabolically related aminophospholipids". The Journal of Lipid Research. 49 (7): 1377–87. doi:10.1194/jlr.R700020-JLR200. PMID 18204094.
- ↑ Deleault, N. R.; Piro, J. R.; Walsh, D. J.; Wang, F.; Ma, J.; Geoghegan, J. C.; Supattapone, S. (2012). "Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids". Proceedings of the National Academy of Sciences. 109 (22): 8546–51. Bibcode:2012PNAS..109.8546D. doi:10.1073/pnas.1204498109. PMC 3365173
 . PMID 22586108.
. PMID 22586108. - ↑ Majumder, R.; Liang, X.; Quinn-Allen, M. A.; Kane, W. H.; Lentz, B. R. (2011). "Modulation of Prothrombinase Assembly and Activity by Phosphatidylethanolamine". Journal of Biological Chemistry. 286 (41): 35535–42. doi:10.1074/jbc.M111.260141. PMC 3195639
 . PMID 21859710.
. PMID 21859710. - ↑ Christie, W.W. (April 16, 2012). "Phosphatidylethanolamine and Related Lipids". The AOCS Lipid Library. Retrieved September 3, 2012.
- ↑ Gbaguidi, B.; Hakizimana, P.; Vandenbussche, G.; Ruysschaert, J.-M. (2007). "Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent". Cellular and Molecular Life Sciences. 64 (12): 1571–82. doi:10.1007/s00018-007-7031-0. PMID 17530171.
- ↑ Kelly, Karen (July 28, 2011). "Phospholipid Biosynthesis". The AOCS Lipid Library. Retrieved September 3, 2012.
- 1 2 Oak, Jeong-Ho; Nakagawa, Kiyotaka; Miyazawa, Teruo (2002). "UV analysis of Amadori-glycated phosphatidylethanolamine in foods and biological samples". The Journal of Lipid Research. 43 (3): 523–9. PMID 11893788.
- ↑ Oak, Jeong-Ho; Nakagawa, Kiyotaka; Miyazawa, Teruo (2000). "Synthetically prepared Amadori-glycated phosphatidylethanolamine can trigger lipid peroxidation via free radical reactions". FEBS Letters. 481 (1): 26–30. doi:10.1016/S0014-5793(00)01966-9. PMID 10984609.
- ↑ Oak, Jeong-Ho; Nakagawa, Kiyotaka; Oikawa, Shinichi; Miyazawa, Teruo (2003). "Amadori-glycated phosphatidylethanolamine induces angiogenic differentiations in cultured human umbilical vein endothelial cells". FEBS Letters. 555 (2): 419–23. doi:10.1016/S0014-5793(03)01237-7. PMID 14644453.
- ↑ Eitsuka, Takahiro; Nakagawa, Kiyotaka; Ono, Yuichi; Tatewaki, Naoto; Nishida, Hiroshi; Kurata, Tadao; Shoji, Naoki; Miyazawa, Teruo (2012). "Amadori-glycated phosphatidylethanolamine up-regulates telomerase activity in PANC-1 human pancreatic carcinoma cells". FEBS Letters. 586 (16): 2542–7. doi:10.1016/j.febslet.2012.06.027. PMID 22750441.
- ↑ Ariizumi, Ken; Koike, T; Ohara, S; Inomata, Y; Abe, Y; Iijima, K; Imatani, A; Oka, T; Shimosegawa, T (2008). "Incidence of reflux esophagitis and H pylori infection in diabetic patients". World Journal of Gastroenterology. 14 (20): 3212–7. doi:10.3748/wjg.14.3212. PMC 2712855
 . PMID 18506928.
. PMID 18506928.
External links
- Phosphatidylethanolamines at the US National Library of Medicine Medical Subject Headings (MeSH)
- Phosphatidylethanolamine at the The AOCS Lipid Library.