Vinculin
| View/Edit Human | View/Edit Mouse |
In mammalian cells, vinculin is a membrane-cytoskeletal protein in focal adhesion plaques that is involved in linkage of integrin adhesion molecules to the actin cytoskeleton. Vinculin is a cytoskeletal protein associated with cell-cell and cell-matrix junctions, where it is thought to function as one of several interacting proteins involved in anchoring F-actin to the membrane.
Discovered independently by Benny Geiger[3] and Keith Burridge,[4] its sequence is 20%-30% similar to α-catenin, which serves a similar function.
Binding alternately to talin or α-actinin, vinculin's shape and, as a consequence, its binding properties are changed. The vinculin gene occurs as a single copy and what appears to be no close relative to take over functions in its absence. Its splice variant metavinculin (see below) also needs vinculin to heterodimerize and work in a dependent fashion.
Structure
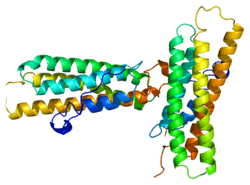
Vinculin is a 117-kDa cytoskeletal protein with 1066 amino acids. The protein contains an acidic N-terminal domain and a basic C-terminal domain separated by a proline-rich middle segment. Vinculin consists of a globular head domain that contains binding sites for talin and α-actinin as well as a tyrosine phosphorylation site, while the tail region contains binding sites for F-actin, paxillin, and lipids.[5]
Essentially, there is an 835 amino acid N-terminal head, which is split into four domains. This is linked to the C-terminal tail with a linker region.
The recent discovery of the 3D structure sheds light on how this protein tailors its shape to perform a variety of functions. For example, vinculin is able to control the cell’s motility by simply altering its shape from active to inactive. When in its ‘inactive’ state, vinculin’s conformation is characterized by the interaction between its head and tail domains. And, when transforming to the ‘active’ form, such as when talin triggers binding, the intramolecular interaction between the tail and head is severed. In other words, when talin’s binding sites (VBS) of α-helices bind to a helical bundle structure in vinculin’s head domain, the ‘helical bundle conversion’ is initiated, which leads to the reorganization of the α-helices (α1- α-4), resulting in an entirely new five-helical bundle structure. This function also extends to cancer cells, and regulating their movement and proliferation of cancer to other parts of the body.
Mechanism and function
Cell spreading and movement occur through the process of binding of cell surface integrin receptors to extracellular matrix adhesion molecules. Vinculin is associated with focal adhesion and adherens junctions, which are complexes that nucleate actin filaments and crosslinkers between the external medium, plasma membrane, and actin cytoskeleton.[6] The complex at the focal adhesions consists of several proteins such as vinculin, α-actin, paxillin, and talin, at the intracellular face of the plasma membrane.
In more specific terms, the amino-terminus of vinculin binds to talin, which, in turn, binds to β-integrins, and the carboxy-terminus binds to actin, phospholipids, and paxillin-forming homodimers. The binding of vinculin to talin and actin is regulated by polyphosphoinositides and inhibited by acidic phospholipids. The complex then serves to anchor actin filaments to the membrane.[7]
The loss of vinculin impacts a variety of cell functions; it disrupts the formation of the complex, and prevents cell adhesion and spreading. The absence of the protein demonstrates a decrease in spreading of cells, accompanied by reduced stress fiber formation, formation of fewer focal adhesions, and inhibition of lamellipodia extension.[5] It was discovered that cells that are deficient in vinculin have growth cones that advance more slowly, as well as filopodia and lamellipoida that were less stable than the wild-type. Based on research, it has been postulated that the lack of vinculin may decrease cell adhesion by inhibiting focal adhesion assembly and preventing actin polymerization. On the other hand, overexpression of vinculin may restore adhesion and spreading by promoting recruitment of cytoskeletal proteins to the focal adhesion complex at the site of integrin binding.[7] Vinculin's ability to interact with integrins to the cytoskeleton at the focal adhesion appears to be critical for control of cytoskeletal mechanics, cell spreading, and lamellipodia formation. Thus, vinculin appears to play a key role in shape control based on its ability to modulate focal adhesion structure and function.
Vinculin binding site
| VBS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
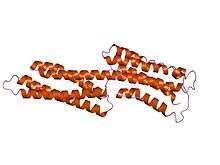 human vinculin head (1-258) in complex with talin's vinculin binding site 3 (residues 1944-1969) | |||||||||
| Identifiers | |||||||||
| Symbol | VBS | ||||||||
| Pfam | PF08913 | ||||||||
| InterPro | IPR015009 | ||||||||
| |||||||||
Vinculin binding sites are predominantly found in talin and talin-like molecules, enabling binding of vinculin to talin, stabilising integrin-mediated cell-matrix junctions. Talin, in turn, links integrins to the actin cytoskeleton. The consensus sequence for Vinculin binding sites is LxxAAxxVAxxVxxLIxxA, with a secondary structure prediction of four amphipathic helices. The hydrophobic residues that define the VBS are themselves 'masked' and are buried in the core of a series of helical bundles that make up the talin rod.[8]
Metavinculin splice variant
Smooth muscles and skeletal muscles (and probably to a lower extent in cardiac muscle) in their well-differentiated (contractile) state co-express (along with vinculin) a splice variant carrying an extra exon in the 3' coding region, thus encoding a longer isoform meta-vinculin (meta VCL) of ~150KD molecular weight — a protein whose existence has been known since the 1980s.[9] Translation of the extra exon causes a 68- to 79-amino acid acid-rich insert between helices I and II within the C-terminal tail domain. Mutations within the insert region correlate with hereditary idiopathic dilated cardiomyopathy.[10]
The length of the insert in metavinculin is 68 AA in mammals and 79 in frog.[11] Compared metavinculin sequences from pig, man, chicken, and frog, and found the insert to be bipartite: the first part variable and the second highly conserved. Both vinculin isoforms co-localize in muscular adhesive structures, such as dense plaques in smooth muscles, intercalated discs in cardiomyocytes, and costameres in skeletal muscles.[12] Metavinculin tail domain has a lower affinity for the head as compared with the vinculin tail. In case of metavinculin, unfurling of the C-terminal hydrophobic hairpin loop of tail domain is impaired by the negative charges of the 68-amino acid insert, thus requiring phospholipid-activated regular isoform of vinculin to fully activate the metavinculin molecule.
Interactions
Vinculin has been shown to interact with:
In cases of Small Intestinal Bacterial Overgrowth presented as IBS symptoms, anti-cdtb antibodies have been identified to affect vinculin function, which is required in gut motility.[19]
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Geiger B (1979). "A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells". Cell. 18 (1): 193–205. doi:10.1016/0092-8674(79)90368-4. PMID 574428.
- ↑ Burridge K, Feramisco JR (1980). "Microinjection and localization of a 130K protein in living fibroblasts: a relationship to actin and fibronectin". Cell. 19 (3): 587–95. doi:10.1016/s0092-8674(80)80035-3. PMID 6988083.
- 1 2 Goldmann WH, Ingber DE (2001). "Intact vinculin protein is Require for control of cell shape, cell mechanics, and rac-Dependent Lamellipodia Formation". Biochemical and Biophysical Research Communications. 290 (2): 749–755. doi:10.1006/bbrc.2001.6243. PMID 11785963.
- ↑ Xu W, Baribault H, Adamson ED (1998). "vinculin knockout results in heart and brain defects during embryonic development". Development. 125 (2): 327–337. PMID 9486805.
- 1 2 Ezzell RM, Goldmann WH, Wang N, Parashurama N, Parasharama N, Ingber DE (1997). "Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton". Exp. Cell Res. 231 (1): 14–26. doi:10.1006/excr.1996.3451. PMID 9056408.
- ↑ Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL (February 2006). "Structural and dynamic characterization of a vinculin binding site in the talin rod". Biochemistry. 45 (6): 1805–17. doi:10.1021/bi052136l. PMID 16460027.
- ↑ Feramisco JR, Smart JE, Burridge K, Helfman DM, Thomas GP (1982). "Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle". J. Biol. Chem. 257 (18): 11024–31. PMID 6809764.
- ↑ Witt S, Zieseniss A, Fock U, Jockusch BM, Illenberger S (2004). "Comparative biochemical analysis suggests that vinculin and metavinculin cooperate in muscular adhesion sites". J. Biol. Chem. 279 (30): 31533–43. doi:10.1074/jbc.M314245200. PMID 15159399.
- ↑ Strasser P, Gimona M, Herzog M, Geiger B, Small JV (1993). "Variable and constant regions in the C-terminus of vinculin and metavinculin. Cloning and expression of fragments in E. coli". FEBS Lett. 317 (3): 189–94. doi:10.1016/0014-5793(93)81274-4. PMID 8425604.
- ↑ Belkin AM, Ornatsky OI, Glukhova MA, Koteliansky VE (1988). "Immunolocalization of meta-vinculin in human smooth and cardiac muscles". J. Cell Biol. 107 (2): 545–53. doi:10.1083/jcb.107.2.545. PMC 2115213
 . PMID 3138246.
. PMID 3138246. - ↑ Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL (December 1997). "Vinculin is associated with the E-cadherin adhesion complex". J. Biol. Chem. 272 (51): 32448–53. doi:10.1074/jbc.272.51.32448. PMID 9405455.
- ↑ Hazan RB, Norton L (April 1998). "The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton". J. Biol. Chem. 273 (15): 9078–84. doi:10.1074/jbc.273.15.9078. PMID 9535896.
- ↑ Turner CE, Brown MC, Perrotta JA, Riedy MC, Nikolopoulos SN, McDonald AR, Bagrodia S, Thomas S, Leventhal PS (May 1999). "Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: A role in cytoskeletal remodeling". J. Cell Biol. 145 (4): 851–63. doi:10.1083/jcb.145.4.851. PMC 2133183
 . PMID 10330411.
. PMID 10330411. - ↑ Mazaki Y, Hashimoto S, Sabe H (March 1997). "Monocyte cells and cancer cells express novel paxillin isoforms with different binding properties to focal adhesion proteins". J. Biol. Chem. 272 (11): 7437–44. doi:10.1074/jbc.272.11.7437. PMID 9054445.
- ↑ Brown MC, Perrotta JA, Turner CE (November 1996). "Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding". J. Cell Biol. 135 (4): 1109–23. doi:10.1083/jcb.135.4.1109. PMC 2133378
 . PMID 8922390.
. PMID 8922390. - ↑ Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y (March 1999). "Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions". J. Cell Biol. 144 (5): 1001–17. doi:10.1083/jcb.144.5.1001. PMC 2148189
 . PMID 10085297.
. PMID 10085297. - ↑ Pimentel M, Morales W, Pokkunuri V, Brikos C, Kim SM, Kim SE, Triantafyllou K, Weitsman S, Marsh Z, Marsh E, Chua KS, Srinivasan S, Barlow GM, Chang C (May 2015). "Autoimmunity Links Vinculin to the Pathophysiology of Chronic Functional Bowel Changes Following Campylobacter jejuni Infection in a Rat Model.". Digestive Diseases and Sciences. 60 (5): 1195–205. doi:10.1007/s10620-014-3435-5. PMID 25424202.
Further reading
- Critchley DR (2005). "Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion". Biochem. Soc. Trans. 32 (Pt 5): 831–6. doi:10.1042/BST0320831. PMID 15494027.
- Koteliansky VE, Ogryzko EP, Zhidkova NI, Weller PA, Critchley DR, Vancompernolle K, Vandekerckhove J, Strasser P, Way M, Gimona M (1992). "An additional exon in the human vinculin gene specifically encodes meta-vinculin-specific difference peptide. Cross-species comparison reveals variable and conserved motifs in the meta-vinculin insert". Eur. J. Biochem. 204 (2): 767–72. doi:10.1111/j.1432-1033.1992.tb16692.x. PMID 1339348.
- Mulligan LM, Gardner E, Telenius H, Ponder BA (1992). "Complementary physical and genetic techniques map the vinculin (VCL) gene on chromosome 10q". Genomics. 13 (4): 1347–9. doi:10.1016/0888-7543(92)90066-2. PMID 1505973.
- Weller PA, Ogryzko EP, Corben EB, Zhidkova NI, Patel B, Price GJ, Spurr NK, Koteliansky VE, Critchley DR (1990). "Complete sequence of human vinculin and assignment of the gene to chromosome 10". Proc. Natl. Acad. Sci. U.S.A. 87 (15): 5667–71. doi:10.1073/pnas.87.15.5667. PMC 54388
 . PMID 2116004.
. PMID 2116004. - Turner CE, Burridge K (1989). "Detection of metavinculin in human platelets using a modified talin overlay assay". Eur. J. Cell Biol. 49 (1): 202–6. PMID 2503380.
- Turner CE, Miller JT (1994). "Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region". J. Cell. Sci. 107 (6): 1583–91. PMID 7525621.
- Salgia R, Li JL, Lo SH, Brunkhorst B, Kansas GS, Sobhany ES, Sun Y, Pisick E, Hallek M, Ernst T (1995). "Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL". J. Biol. Chem. 270 (10): 5039–47. doi:10.1074/jbc.270.10.5039. PMID 7534286.
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O (1995). "Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence" (PDF). Nature. 377 (6547 Suppl): 3–174. PMID 7566098.
- Hagmann J (1993). "Pattern formation and handedness in the cytoskeleton of human platelets". Proc. Natl. Acad. Sci. U.S.A. 90 (8): 3280–3. doi:10.1073/pnas.90.8.3280. PMC 46283
 . PMID 7682697.
. PMID 7682697. - Johnson RP, Craig SW (1995). "F-actin binding site masked by the intramolecular association of vinculin head and tail domains". Nature. 373 (6511): 261–4. doi:10.1038/373261a0. PMID 7816144.
- Hirsch MS, Law LY, Trinkaus-Randall V, Svoboda KK (1995). "The intracellular distribution of vinculin and alpha 2 integrin in epithelial cells and chondrocytes". Scanning. 16 (5): 275–84. doi:10.1002/sca.4950160306. PMID 7994488.
- Fausser JL, Ungewickell E, Ruch JV, Lesot H (1994). "Interaction of vinculin with the clathrin heavy chain". J. Biochem. 114 (4): 498–503. PMID 8276759.
- Moiseyeva EP, Weller PA, Zhidkova NI, Corben EB, Patel B, Jasinska I, Koteliansky VE, Critchley DR (1993). "Organization of the human gene encoding the cytoskeletal protein vinculin and the sequence of the vinculin promoter". J. Biol. Chem. 268 (6): 4318–25. PMID 8440716.
- Yoshida M, Westlin WF, Wang N, Ingber DE, Rosenzweig A, Resnick N, Gimbrone MA (1996). "Leukocyte adhesion to vascular endothelium induces E-selectin linkage to the actin cytoskeleton". J. Cell Biol. 133 (2): 445–55. doi:10.1083/jcb.133.2.445. PMC 2120789
 . PMID 8609175.
. PMID 8609175. - Scott GA, Liang H, Cassidy LL (1996). "Developmental regulation of focal contact protein expression in human melanocytes". Pigment Cell Res. 8 (4): 221–8. doi:10.1111/j.1600-0749.1995.tb00667.x. PMID 8610074.
- Deroanne CF, Colige AC, Nusgens BV, Lapiere CM (1996). "Modulation of expression and assembly of vinculin during in vitro fibrillar collagen-induced angiogenesis and its reversal". Exp. Cell Res. 224 (2): 215–23. doi:10.1006/excr.1996.0131. PMID 8612698.
- Maeda M, Holder E, Lowes B, Valent S, Bies RD (1997). "Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin". Circulation. 95 (1): 17–20. doi:10.1161/01.cir.95.1.17. PMID 8994410.
- Mazaki Y, Hashimoto S, Sabe H (1997). "Monocyte cells and cancer cells express novel paxillin isoforms with different binding properties to focal adhesion proteins". J. Biol. Chem. 272 (11): 7437–44. doi:10.1074/jbc.272.11.7437. PMID 9054445.
- Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL (1998). "Vinculin is associated with the E-cadherin adhesion complex". J. Biol. Chem. 272 (51): 32448–53. doi:10.1074/jbc.272.51.32448. PMID 9405455.
- Hazan RB, Norton L (1998). "The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton". J. Biol. Chem. 273 (15): 9078–84. doi:10.1074/jbc.273.15.9078. PMID 9535896.
External links
- MBInfo - Vinculin
- http://www.stjude.org/media/0,2561,453_5297_9660,00.html
- http://www.medicalnewstoday.com/medicalnews.php?newsid=5196
- http://www.olympusmicro.com/primer/techniques/fluorescence/gallery/cells/bpae/images/bpaelarge.jpg
- Vinculin Info with links in the Cell Migration Gateway