Archaea
| Archaea (Archaebacteria) Temporal range: Paleoarchean or perhaps Eoarchean – Recent | |
|---|---|
 | |
| Halobacteria sp. strain NRC-1, each cell about 5 μm long | |
| Scientific classification | |
| Superdomain: | Neomura |
| Domain: | Archaea Woese, Kandler & Wheelis, 1990[1] |
| Kingdoms[2] and phyla[3] | |
| |
| Synonyms | |
| |
The Archaea (![]() i/ɑːrˈkiːə/ or /ɑːrˈkeɪə/ ar-KEE-ə or ar-KAY-ə) constitute a domain and kingdom of single-celled microorganisms. These microbes (archaea; singular archaeon) are prokaryotes, meaning that they have no cell nucleus or any other membrane-bound organelles in their cells.
i/ɑːrˈkiːə/ or /ɑːrˈkeɪə/ ar-KEE-ə or ar-KAY-ə) constitute a domain and kingdom of single-celled microorganisms. These microbes (archaea; singular archaeon) are prokaryotes, meaning that they have no cell nucleus or any other membrane-bound organelles in their cells.
Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this classification is outdated.[4] Archaeal cells have unique properties separating them from the other two domains of life, Bacteria and Eukaryota. The Archaea are further divided into multiple recognized phyla. Classification is difficult because the majority have not been isolated in the laboratory and have only been detected by analysis of their nucleic acids in samples from their environment.
Archaea and bacteria are generally similar in size and shape, although a few archaea have very strange shapes, such as the flat and square-shaped cells of Haloquadratum walsbyi.[5] Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably the enzymes involved in transcription and translation. Other aspects of archaeal biochemistry are unique, such as their reliance on ether lipids in their cell membranes, including archaeols. Archaea use more energy sources than eukaryotes: these range from organic compounds, such as sugars, to ammonia, metal ions or even hydrogen gas. Salt-tolerant archaea (the Haloarchaea) use sunlight as an energy source, and other species of archaea fix carbon; however, unlike plants and cyanobacteria, no known species of archaea does both. Archaea reproduce asexually by binary fission, fragmentation, or budding; unlike bacteria and eukaryotes, no known species forms spores.
Archaea were initially viewed as extremophiles living in harsh environments, such as hot springs and salt lakes, but they have since been found in a broad range of habitats, including soils, oceans, marshlands and the human colon, oral cavity, and skin.[6] Archaea are particularly numerous in the oceans, and the archaea in plankton may be one of the most abundant groups of organisms on the planet. Archaea are a major part of Earth's life and may play roles in both the carbon cycle and the nitrogen cycle. No clear examples of archaeal pathogens or parasites are known, but they are often mutualists or commensals. One example is the methanogens that inhabit human and ruminant guts, where their vast numbers aid digestion. Methanogens are also used in biogas production and sewage treatment, and enzymes from extremophile archaea that can endure high temperatures and organic solvents are exploited in biotechnology.
Classification
New domain
For much of the 20th century, prokaryotes were regarded as a single group of organisms and classified based on their biochemistry, morphology and metabolism. For example, microbiologists tried to classify microorganisms based on the structures of their cell walls, their shapes, and the substances they consume.[7] In 1965, Emile Zuckerkandl and Linus Pauling[8] proposed instead using the sequences of the genes in different prokaryotes to work out how they are related to each other. This approach, known as phylogenetics, is the main method used today.
Archaea were first classified as a separate group of prokaryotes in 1977 by Carl Woese and George E. Fox in phylogenetic trees based on the sequences of ribosomal RNA (rRNA) genes.[9] These two groups were originally named the Archaebacteria and Eubacteria and treated as kingdoms or subkingdoms, which Woese and Fox termed Urkingdoms. Woese argued that this group of prokaryotes is a fundamentally different sort of life. To emphasize this difference, Woese later proposed a new natural system of organisms with three separate Domains: the Eukarya, the Bacteria and the Archaea,[1] in what is now known as "The Woesian Revolution".
The word archaea comes from the Ancient Greek ἀρχαῖα, meaning "ancient things",[10] as the first representatives of the domain Archaea were methanogens and it was assumed that their metabolism reflected Earth's primitive atmosphere and the organism's antiquity. For a long time, archaea were seen as extremophiles that only exist in extreme habitats such as hot springs and salt lakes. However, as new habitats were studied, more organisms were discovered. Extreme halophilic[11] and hyperthermophilic microbes[12] were also included in the Archaea. By the end of the 20th century, archaea had been identified in non-extreme environments as well. Today, they are known to be a large and diverse group of organisms that are widely distributed in nature and are common in all habitats.[13] This new appreciation of the importance and ubiquity of archaea came from using polymerase chain reaction (PCR) to detect prokaryotes from environmental samples (such as water or soil) by multiplying their ribosomal genes. This allows the detection and identification of organisms that have not been cultured in the laboratory.[14][15]
Current classification

The classification of archaea, and of prokaryotes in general, is a rapidly moving and contentious field. Current classification systems aim to organize archaea into groups of organisms that share structural features and common ancestors.[16] These classifications rely heavily on the use of the sequence of ribosomal RNA genes to reveal relationships between organisms (molecular phylogenetics).[17] Most of the culturable and well-investigated species of archaea are members of two main phyla, the Euryarchaeota and Crenarchaeota. Other groups have been tentatively created. For example, the peculiar species Nanoarchaeum equitans, which was discovered in 2003, has been given its own phylum, the Nanoarchaeota.[18] A new phylum Korarchaeota has also been proposed. It contains a small group of unusual thermophilic species that shares features of both of the main phyla, but is most closely related to the Crenarchaeota.[19][20] Other recently detected species of archaea are only distantly related to any of these groups, such as the Archaeal Richmond Mine acidophilic nanoorganisms (ARMAN), which were discovered in 2006[21] and are some of the smallest organisms known.[22]
A superphylum - TACK - has been proposed that includes the Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota.[23] This superphylum may be related to the origin of eukaryotes.
Concept of species
The classification of archaea into species is also controversial. Biology defines a species as a group of related organisms. The familiar exclusive breeding criterion (organisms that can breed with each other but not with others) is of no help here because archaea reproduce asexually.[24]
Archaea show high levels of horizontal gene transfer between lineages. Some researchers suggest that individuals can be grouped into species-like populations given highly similar genomes and infrequent gene transfer to/from cells with less-related genomes, as in the genus Ferroplasma.[25] On the other hand, studies in Halorubrum found significant genetic transfer to/from less-related populations, limiting the criterion's applicability.[26] A second concern is to what extent such species designations have practical meaning.[27]
Current knowledge on genetic diversity is fragmentary and the total number of archaeal species cannot be estimated with any accuracy.[17] Estimates of the number of phyla range from 18 to 23, of which only 8 have representatives that have been cultured and studied directly. Many of these hypothesized groups are known from a single rRNA sequence, indicating that the diversity among these organisms remains obscure.[28] The Bacteria also contain many uncultured microbes with similar implications for characterization.[29]
Origin and evolution
The age of the Earth is about 4.54 billion years old.[30][31][32] Scientific evidence suggests that life began on Earth at least 3.5 billion years ago.[33][34] The earliest evidence for life on Earth is graphite found to be biogenic in 3.7 billion-year-old metasedimentary rocks discovered in Western Greenland[35] and microbial mat fossils found in 3.48 billion-year-old sandstone discovered in Western Australia.[36][37] More recently, in 2015, "remains of biotic life" were found in 4.1 billion-year-old rocks in Western Australia.[38][39] An unrelated researcher, S. Blair Hedges, wrote in an email about the study saying, "If life arose relatively quickly on Earth ... then it could be common in the universe."[38]
Although probable prokaryotic cell fossils date to almost 3.5 billion years ago, most prokaryotes do not have distinctive morphologies and fossil shapes cannot be used to identify them as archaea.[40] Instead, chemical fossils of unique lipids are more informative because such compounds do not occur in other organisms.[41] Some publications suggest that archaeal or eukaryotic lipid remains are present in shales dating from 2.7 billion years ago;[42] such data have since been questioned.[43] Such lipids have also been detected in even older rocks from west Greenland. The oldest such traces come from the Isua district, which include Earth's oldest known sediments, formed 3.8 billion years ago.[44] The archaeal lineage may be the most ancient that exists on Earth.[45]
Woese argued that the bacteria, archaea, and eukaryotes represent separate lines of descent that diverged early on from an ancestral colony of organisms.[46][47] One possibility[47][48] is that this occurred before the evolution of cells, when the lack of a typical cell membrane allowed unrestricted lateral gene transfer, and that the common ancestors of the three domains arose by fixation of specific subsets of genes.[47][48] It is possible that the last common ancestor of the bacteria and archaea was a thermophile, which raises the possibility that lower temperatures are "extreme environments" in archaeal terms, and organisms that live in cooler environments appeared only later.[49] Since the Archaea and Bacteria are no more related to each other than they are to eukaryotes, the term prokaryote's only surviving meaning is "not a eukaryote", limiting its value.[50]
Comparison to other domains
The following table compares some major characteristics of the three domains, to illustrate their similarities and differences.[51] Many of these characteristics are also discussed below.
| Property | Archaea | Bacteria | Eukarya |
|---|---|---|---|
| Cell Membrane | Ether-linked lipids, pseudopeptidoglycan | Ester-linked lipids, peptidoglycan | Ester-linked lipids, various structures |
| Gene Structure | Circular chromosomes, similar translation and transcription to Eukarya | Circular chromosomes, unique translation and transcription | Multiple, linear chromosomes, similar translation and transcription to Archaea |
| Internal Cell Structure | No membrane-bound organelles or nucleus | No membrane-bound organelles or nucleus | Membrane-bound organelles and nucleus |
| Metabolism [52] | Various, with methanogenesis unique to Archaea | Various, including photosynthesis, aerobic and anaerobic respiration, fermentation, and autotrophy | Photosynthesis and cellular respiration |
| Reproduction | Asexual reproduction, horizontal gene transfer | Asexual reproduction, horizontal gene transfer | Sexual and asexual reproduction |
Archaea were split off as a third domain because of the large differences in their ribosomal RNA structure. The particular RNA molecule sequenced, known as 16s rRNA, is present in all organisms and always has the same vital function, the production of proteins. Because this function is so central to life, organisms with mutations of its 16s rRNA are unlikely to survive, leading to great stability in the structure of this nucleotide over many generations. 16s rRNA is also large enough to retain organism-specific information, but small enough to be sequenced in a manageable amount of time. In 1977, Carl Woese, a microbiologist studying the genetic sequencing of organisms, developed a new sequencing method that involved splitting the RNA into fragments that could be sorted and compared to other fragments from other organisms.[53] The more similar the patterns between species were, the more closely related the organisms.[54]
Woese used his new rRNA comparison method to categorize and contrast different organisms. He sequenced a variety of different species and happened upon a group of methanogens that had vastly different patterns than any known prokaryotes or eukaryotes.[53] These methanogens were much more similar to each other than they were to other organisms sequenced, leading Woese to propose the new domain of Archaea.[53] One of the interesting results of his experiments was that the Archaea were more similar to eukaryotes than prokaryotes, even though they were more similar to prokaryotes in structure.[55] This led to the conclusion that Archaea and Eukarya shared a more recent common ancestor than Eukarya and Bacteria in general.[55] The development of the nucleus occurred after the split between Bacteria and this common ancestor.[55] Although Archaea are prokaryotic, they are more closely related to Eukarya and thus cannot be placed within either the Bacteria or Eukarya domains.[1]
One property unique to Archaea is the abundant use of ether-linked lipids in their cell membranes. Ether linkages are more chemically stable than the ester linkages found in Bacteria and Eukarya, which may be a contributing factor to the ability of many Archaea to survive in extreme environments that place heavy stress on cell membranes, such as extreme heat and salinity. Another unique feature of Archaea is that no other known organisms are capable of methanogenesis (the metabolic production of methane). Methanogenic Archaea play a pivotal role in ecosystems with organisms that derive energy from oxidation of methane, many of which are Bacteria, as they are often a major source of methane in such environments and can play a role as primary producers. Methanogens also play a critical role in the carbon cycle, breaking down organic carbon into methane, which is also a major greenhouse gas.[56]
Relationship to other prokaryotes
Error: No valid link was found at the end of line 3.
The relationship between the three domains is of central importance for understanding the origin of life. Most of the metabolic pathways, which comprise the majority of an organism's genes, are common between Archaea and Bacteria, while most genes involved in genome expression are common between Archaea and Eukarya.[57] Within prokaryotes, archaeal cell structure is most similar to that of gram-positive bacteria, largely because both have a single lipid bilayer[58] and usually contain a thick sacculus (exoskeleton) of varying chemical composition.[59] In some phylogenetic trees based upon different gene/protein sequences of prokaryotic homologs, the archaeal homologs are more closely related to those of gram-positive bacteria.[58] Archaea and gram-positive bacteria also share conserved indels in a number of important proteins, such as Hsp70 and glutamine synthetase I;[58][60] however, the phylogeny of these genes was interpreted to reveal interdomain gene transfer,[61][62] and might not reflect the organismal relationship(s).
It has been proposed that the archaea evolved from gram-positive bacteria in response to antibiotic selection pressure.[58][60][63] This is suggested by the observation that archaea are resistant to a wide variety of antibiotics that are primarily produced by gram-positive bacteria,[58][60] and that these antibiotics primarily act on the genes that distinguish archaea from bacteria. The proposal is that the selective pressure towards resistance generated by the gram-positive antibiotics was eventually sufficient to cause extensive changes in many of the antibiotics' target genes, and that these strains represented the common ancestors of present-day Archaea.[63] The evolution of Archaea in response to antibiotic selection, or any other competitive selective pressure, could also explain their adaptation to extreme environments (such as high temperature or acidity) as the result of a search for unoccupied niches to escape from antibiotic-producing organisms;[63][64] Cavalier-Smith has made a similar suggestion.[65] This proposal is also supported by other work investigating protein structural relationships[66] and studies that suggest that gram-positive bacteria may constitute the earliest branching lineages within the prokaryotes.[67]
Relation to eukaryotes
The evolutionary relationship between archaea and eukaryotes remains unclear. Aside from the similarities in cell structure and function that are discussed below, many genetic trees group the two.
Complicating factors include claims that the relationship between eukaryotes and the archaeal phylum Crenarchaeota is closer than the relationship between the Euryarchaeota and the phylum Crenarchaeota[68] and the presence of archaea-like genes in certain bacteria, such as Thermotoga maritima, from horizontal gene transfer.[69] The standard hypothesis states that the ancestor of the eukaryotes diverged early from the Archaea,[70][71] and that eukaryotes arose through fusion of an archaean and eubacterium, which became the nucleus and cytoplasm; this explains various genetic similarities but runs into difficulties explaining cell structure.[72] An alternative hypothesis, the eocyte hypothesis, posits that Eukaryota emerged relatively late from the Archaea.[73]
A recently discovered lineage of archaea, Lokiarchaeum, named for a hydrothermal vent called Loki's Castle in the Arctic Ocean, has been found to be most closely related to eukaryotes. It has been called a transitional organism between prokaryotes and eukaryotes.[74][75]
Morphology
Individual archaea range from 0.1 micrometers (μm) to over 15 μm in diameter, and occur in various shapes, commonly as spheres, rods, spirals or plates.[76] Other morphologies in the Crenarchaeota include irregularly shaped lobed cells in Sulfolobus, needle-like filaments that are less than half a micrometer in diameter in Thermofilum, and almost perfectly rectangular rods in Thermoproteus and Pyrobaculum.[77] Haloquadratum walsbyi are flat, square archaea that live in hypersaline pools.[78] These unusual shapes are probably maintained both by their cell walls and a prokaryotic cytoskeleton. Proteins related to the cytoskeleton components of other organisms exist in archaea,[79] and filaments form within their cells,[80] but in contrast to other organisms, these cellular structures are poorly understood.[81] In Thermoplasma and Ferroplasma the lack of a cell wall means that the cells have irregular shapes, and can resemble amoebae.[82]
Some species form aggregates or filaments of cells up to 200 μm long.[76] These organisms can be prominent in biofilms.[83] Notably, aggregates of Thermococcus coalescens cells fuse together in culture, forming single giant cells.[84] Archaea in the genus Pyrodictium produce an elaborate multicell colony involving arrays of long, thin hollow tubes called cannulae that stick out from the cells' surfaces and connect them into a dense bush-like agglomeration.[85] The function of these cannulae is not settled, but they may allow communication or nutrient exchange with neighbors.[86] Multi-species colonies exist, such as the "string-of-pearls" community that was discovered in 2001 in a German swamp. Round whitish colonies of a novel Euryarchaeota species are spaced along thin filaments that can range up to 15 centimetres (5.9 in) long; these filaments are made of a particular bacteria species.[87]
Structure, composition development, and operation
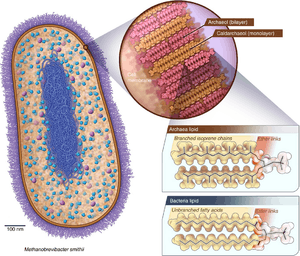
Archaea and bacteria have generally similar cell structure, but cell composition and organization set the archaea apart. Like bacteria, archaea lack interior membranes and organelles.[50] Like bacteria, archaea cell membranes are usually bounded by a cell wall and they swim using one or more flagella.[88] Structurally, archaea are most similar to gram-positive bacteria. Most have a single plasma membrane and cell wall, and lack a periplasmic space; the exception to this general rule is Ignicoccus, which possess a particularly large periplasm that contains membrane-bound vesicles and is enclosed by an outer membrane.[89]
Cell wall and flagella
Most archaea (but not Thermoplasma and Ferroplasma) possess a cell wall.[82] In most archaea the wall is assembled from surface-layer proteins, which form an S-layer.[90] An S-layer is a rigid array of protein molecules that cover the outside of the cell (like chain mail).[91] This layer provides both chemical and physical protection, and can prevent macromolecules from contacting the cell membrane.[92] Unlike bacteria, archaea lack peptidoglycan in their cell walls.[93] Methanobacteriales do have cell walls containing pseudopeptidoglycan, which resembles eubacterial peptidoglycan in morphology, function, and physical structure, but pseudopeptidoglycan is distinct in chemical structure; it lacks D-amino acids and N-acetylmuramic acid.[92]
Archaea flagella operate like bacterial flagella—their long stalks are driven by rotatory motors at the base. These motors are powered by the proton gradient across the membrane. However, archaeal flagella are notably different in composition and development.[88] The two types of flagella evolved from different ancestors. The bacterial flagellum shares a common ancestor with the type III secretion system,[94][95] while archaeal flagella appear to have evolved from bacterial type IV pili.[96] In contrast to the bacterial flagellum, which is hollow and is assembled by subunits moving up the central pore to the tip of the flagella, archaeal flagella are synthesized by adding subunits at the base.[97]
Membranes
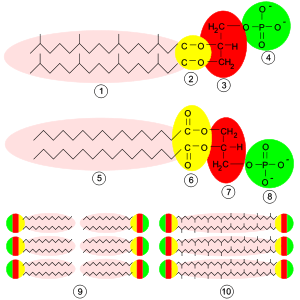
Archaeal membranes are made of molecules that differ strongly from those in other life forms, showing that archaea are related only distantly to bacteria and eukaryotes.[98] In all organisms, cell membranes are made of molecules known as phospholipids. These molecules possess both a polar part that dissolves in water (the phosphate "head"), and a "greasy" non-polar part that does not (the lipid tail). These dissimilar parts are connected by a glycerol moiety. In water, phospholipids cluster, with the heads facing the water and the tails facing away from it. The major structure in cell membranes is a double layer of these phospholipids, which is called a lipid bilayer.
These phospholipids are unusual in four ways:
- Bacteria and eukaryotes have membranes composed mainly of glycerol-ester lipids, whereas archaea have membranes composed of glycerol-ether lipids.[99] The difference is the type of bond that joins the lipids to the glycerol moiety; the two types are shown in yellow in the figure at the right. In ester lipids this is an ester bond, whereas in ether lipids this is an ether bond. Ether bonds are chemically more resistant than ester bonds. This stability might help archaea to survive extreme temperatures and very acidic or alkaline environments.[100] Bacteria and eukaryotes do contain some ether lipids, but in contrast to archaea these lipids are not a major part of their membranes.
- The stereochemistry of the glycerol moiety is the reverse of that found in other organisms. The glycerol moiety can occur in two forms that are mirror images of one another, called the right-handed and left-handed forms; in chemistry these are called enantiomers. Just as a right hand does not fit easily into a left-handed glove, a right-handed phospholipid generally cannot be used or made by enzymes adapted for the left-handed form. This suggests that archaea use entirely different enzymes for synthesizing phospholipids than do bacteria and eukaryotes. Such enzymes developed very early in life's history, suggesting an early split from the other two domains.[98]
- Archaeal lipid tails are chemically different from other organisms. Archaeal lipids are based upon the isoprenoid sidechain and are long chains with multiple side-branches and sometimes even cyclopropane or cyclohexane rings.[101] This is in contrast to the fatty acids found in other organisms' membranes, which have straight chains with no branches or rings. Although isoprenoids play an important role in the biochemistry of many organisms, only the archaea use them to make phospholipids. These branched chains may help prevent archaeal membranes from leaking at high temperatures.[102]
- In some archaea, the lipid bilayer is replaced by a monolayer. In effect, the archaea fuse the tails of two independent phospholipid molecules into a single molecule with two polar heads (a bolaamphiphile); this fusion may make their membranes more rigid and better able to resist harsh environments.[103] For example, the lipids in Ferroplasma are of this type, which is thought to aid this organism's survival in its highly acidic habitat.[104]
Metabolism
Archaea exhibit a great variety of chemical reactions in their metabolism and use many sources of energy. These reactions are classified into nutritional groups, depending on energy and carbon sources. Some archaea obtain energy from inorganic compounds such as sulfur or ammonia (they are lithotrophs). These include nitrifiers, methanogens and anaerobic methane oxidisers.[105] In these reactions one compound passes electrons to another (in a redox reaction), releasing energy to fuel the cell's activities. One compound acts as an electron donor and one as an electron acceptor. The energy released generates adenosine triphosphate (ATP) through chemiosmosis, in the same basic process that happens in the mitochondrion of eukaryotic cells.[106]
Other groups of archaea use sunlight as a source of energy (they are phototrophs). However, oxygen–generating photosynthesis does not occur in any of these organisms.[106] Many basic metabolic pathways are shared between all forms of life; for example, archaea use a modified form of glycolysis (the Entner–Doudoroff pathway) and either a complete or partial citric acid cycle.[107] These similarities to other organisms probably reflect both early origins in the history of life and their high level of efficiency.[108]
| Nutritional type | Source of energy | Source of carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds | Halobacterium |
| Lithotrophs | Inorganic compounds | Organic compounds or carbon fixation | Ferroglobus, Methanobacteria or Pyrolobus |
| Organotrophs | Organic compounds | Organic compounds or carbon fixation | Pyrococcus, Sulfolobus or Methanosarcinales |
Some Euryarchaeota are methanogens living in anaerobic environments, such as swamps. This form of metabolism evolved early, and it is even possible that the first free-living organism was a methanogen.[109] A common reaction involves the use of carbon dioxide as an electron acceptor to oxidize hydrogen. Methanogenesis involves a range of coenzymes that are unique to these archaea, such as coenzyme M and methanofuran.[110] Other organic compounds such as alcohols, acetic acid or formic acid are used as alternative electron acceptors by methanogens. These reactions are common in gut-dwelling archaea. Acetic acid is also broken down into methane and carbon dioxide directly, by acetotrophic archaea. These acetotrophs are archaea in the order Methanosarcinales, and are a major part of the communities of microorganisms that produce biogas.[111]
Other archaea use CO
2 in the atmosphere as a source of carbon, in a process called carbon fixation (they are autotrophs). This process involves either a highly modified form of the Calvin cycle[113] or a recently discovered metabolic pathway called the 3-hydroxypropionate/4-hydroxybutyrate cycle.[114] The Crenarchaeota also use the reverse Krebs cycle while the Euryarchaeota also use the reductive acetyl-CoA pathway.[115] Carbon–fixation is powered by inorganic energy sources. No known archaea carry out photosynthesis.[116] Archaeal energy sources are extremely diverse, and range from the oxidation of ammonia by the Nitrosopumilales[117][118] to the oxidation of hydrogen sulfide or elemental sulfur by species of Sulfolobus, using either oxygen or metal ions as electron acceptors.[106]
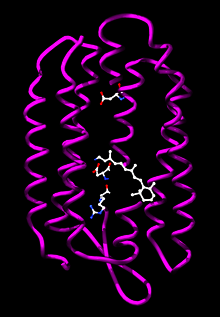
Phototrophic archaea use light to produce chemical energy in the form of ATP. In the Halobacteria, light-activated ion pumps like bacteriorhodopsin and halorhodopsin generate ion gradients by pumping ions out of the cell across the plasma membrane. The energy stored in these electrochemical gradients is then converted into ATP by ATP synthase.[76] This process is a form of photophosphorylation. The ability of these light-driven pumps to move ions across membranes depends on light-driven changes in the structure of a retinol cofactor buried in the center of the protein.[119]
Genetics
Archaea usually have a single circular chromosome,[120] the size of which may be as great as 5,751,492 base pairs in Methanosarcina acetivorans,[121] the largest known archaeal genome. The tiny 490,885 base-pair genome of Nanoarchaeum equitans is one-tenth of this size and the smallest archaeal genome known; it is estimated to contain only 537 protein-encoding genes.[122] Smaller independent pieces of DNA, called plasmids, are also found in archaea. Plasmids may be transferred between cells by physical contact, in a process that may be similar to bacterial conjugation.[123][124]
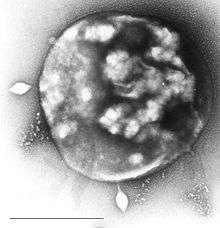
Archaea can be infected by double-stranded DNA viruses that are unrelated to any other form of virus and have a variety of unusual shapes, including bottles, hooked rods, or teardrops.[126] These viruses have been studied in most detail in thermophilics, particularly the orders Sulfolobales and Thermoproteales.[127] Two groups of single-stranded DNA viruses that infect archaea have been recently isolated. One group is exemplified by the Halorubrum pleomorphic virus 1 ("Pleolipoviridae") infecting halophilic archaea[128] and the other one by the Aeropyrum coil-shaped virus ("Spiraviridae") infecting a hyperthermophilic (optimal growth at 90–95 °C) host.[129] Notably, the latter virus has the largest currently reported ssDNA genome. Defenses against these viruses may involve RNA interference from repetitive DNA sequences that are related to the genes of the viruses.[130][131]
Archaea are genetically distinct from bacteria and eukaryotes, with up to 15% of the proteins encoded by any one archaeal genome being unique to the domain, although most of these unique genes have no known function.[132] Of the remainder of the unique proteins that have an identified function, most belong to the Euryarchaea and are involved in methanogenesis. The proteins that archaea, bacteria and eukaryotes share form a common core of cell function, relating mostly to transcription, translation, and nucleotide metabolism.[133] Other characteristic archaeal features are the organization of genes of related function—such as enzymes that catalyze steps in the same metabolic pathway into novel operons, and large differences in tRNA genes and their aminoacyl tRNA synthetases.[133]
Transcription in archaea more closely resembles eukaryotic than bacterial transcription, with the archaeal RNA polymerase being very close to its equivalent in eukaryotes;[120] while archaeal translation shows signs of both bacterial and eukaryal equivalents.[134] Although archaea only have one type of RNA polymerase, its structure and function in transcription seems to be close to that of the eukaryotic RNA polymerase II, with similar protein assemblies (the general transcription factors) directing the binding of the RNA polymerase to a gene's promoter.[135] However, other archaeal transcription factors are closer to those found in bacteria.[136] Post-transcriptional modification is simpler than in eukaryotes, since most archaeal genes lack introns, although there are many introns in their transfer RNA and ribosomal RNA genes,[137] and introns may occur in a few protein-encoding genes.[138][139]
Gene transfer and genetic exchange
Halobacterium volcanii, an extreme halophilic archaeon, forms cytoplasmic bridges between cells that appear to be used for transfer of DNA from one cell to another in either direction.[140]
When the hyperthermophilic archaea Sulfolobus solfataricus[141] and Sulfolobus acidocaldarius[142] are exposed to the DNA damaging agents UV irradiation, bleomycin or mitomycin C, species-specific cellular aggregation is induced. Aggregation in S. solfataricus could not be induced by other physical stressors, such as pH or temperature shift,[141] suggesting that aggregation is induced specifically by DNA damage. Ajon et al.[142] showed that UV-induced cellular aggregation mediates chromosomal marker exchange with high frequency in S. acidocaldarius. Recombination rates exceeded those of uninduced cultures by up to three orders of magnitude. Frols et al.[141][143] and Ajon et al.[142] hypothesized that cellular aggregation enhances species specific DNA transfer between Sulfolobus cells in order to provide increased repair of damaged DNA by means of homologous recombination. This response may be a primitive form of sexual interaction similar to the more well-studied bacterial transformation systems that are also associated with species specific DNA transfer between cells leading to homologous recombinational repair of DNA damage.[144]
Reproduction
Archaea reproduce asexually by binary or multiple fission, fragmentation, or budding; meiosis does not occur, so if a species of archaea exists in more than one form, all have the same genetic material.[76] Cell division is controlled in a cell cycle; after the cell's chromosome is replicated and the two daughter chromosomes separate, the cell divides.[145] In the genus Sulfolobus, the cycle has characteristics that are similar to both bacterial and eukaryotic systems. The chromosomes replicate from multiple starting-points (origins of replication) using DNA polymerases that resemble the equivalent eukaryotic enzymes.[146]
In euryarchaea the cell division protein FtsZ, which forms a contracting ring around the cell, and the components of the septum that is constructed across the center of the cell, are similar to their bacterial equivalents.[145] In cren-[147][148] and thaumarchaea,[149] however, the cell division machinery Cdv fulfills a similar role. This machinery is related to the eukaryotic ESCRT-III machinery which, while best known for its role in cell sorting, also has been seen to fulfill a role in separation between divided cell, suggesting an ancestral role in cell division.
Both bacteria and eukaryotes, but not archaea, make spores.[150] Some species of Haloarchaea undergo phenotypic switching and grow as several different cell types, including thick-walled structures that are resistant to osmotic shock and allow the archaea to survive in water at low salt concentrations, but these are not reproductive structures and may instead help them reach new habitats.[151]
Ecology
Habitats

Archaea exist in a broad range of habitats, and as a major part of global ecosystems,[13] may represent about 20% of microbial cells in the oceans.[152] The first-discovered archaeans were extremophiles.[105] Indeed, some archaea survive high temperatures, often above 100 °C (212 °F), as found in geysers, black smokers, and oil wells. Other common habitats include very cold habitats and highly saline, acidic, or alkaline water. However, archaea include mesophiles that grow in mild conditions, in swamps and marshland, sewage, the oceans, the intestinal tract of animals, and soils.[13]
Extremophile archaea are members of four main physiological groups. These are the halophiles, thermophiles, alkaliphiles, and acidophiles.[153] These groups are not comprehensive or phylum-specific, nor are they mutually exclusive, since some archaea belong to several groups. Nonetheless, they are a useful starting point for classification.
Halophiles, including the genus Halobacterium, live in extremely saline environments such as salt lakes and outnumber their bacterial counterparts at salinities greater than 20–25%.[105] Thermophiles grow best at temperatures above 45 °C (113 °F), in places such as hot springs; hyperthermophilic archaea grow optimally at temperatures greater than 80 °C (176 °F).[154] The archaeal Methanopyrus kandleri Strain 116 can even reproduce at 122 °C (252 °F), the highest recorded temperature of any organism.[155]
Other archaea exist in very acidic or alkaline conditions.[153] For example, one of the most extreme archaean acidophiles is Picrophilus torridus, which grows at pH 0, which is equivalent to thriving in 1.2 molar sulfuric acid.[156]
This resistance to extreme environments has made archaea the focus of speculation about the possible properties of extraterrestrial life.[157] Some extremophile habitats are not dissimilar to those on Mars,[158] leading to the suggestion that viable microbes could be transferred between planets in meteorites.[159]
Recently, several studies have shown that archaea exist not only in mesophilic and thermophilic environments but are also present, sometimes in high numbers, at low temperatures as well. For example, archaea are common in cold oceanic environments such as polar seas.[160] Even more significant are the large numbers of archaea found throughout the world's oceans in non-extreme habitats among the plankton community (as part of the picoplankton).[161] Although these archaea can be present in extremely high numbers (up to 40% of the microbial biomass), almost none of these species have been isolated and studied in pure culture.[162] Consequently, our understanding of the role of archaea in ocean ecology is rudimentary, so their full influence on global biogeochemical cycles remains largely unexplored.[163] Some marine Crenarchaeota are capable of nitrification, suggesting these organisms may affect the oceanic nitrogen cycle,[164] although these oceanic Crenarchaeota may also use other sources of energy.[165] Vast numbers of archaea are also found in the sediments that cover the sea floor, with these organisms making up the majority of living cells at depths over 1 meter below the ocean bottom.[166][167]
Role in chemical cycling
Archaea recycle elements such as carbon, nitrogen and sulfur through their various habitats. Although these activities are vital for normal ecosystem function, archaea can also contribute to human-made changes, and even cause pollution.
Archaea carry out many steps in the nitrogen cycle. This includes both reactions that remove nitrogen from ecosystems (such as nitrate-based respiration and denitrification) as well as processes that introduce nitrogen (such as nitrate assimilation and nitrogen fixation).[168][169] Researchers recently discovered archaeal involvement in ammonia oxidation reactions. These reactions are particularly important in the oceans.[118][170] The archaea also appear crucial for ammonia oxidation in soils. They produce nitrite, which other microbes then oxidize to nitrate. Plants and other organisms consume the latter.[171]
In the sulfur cycle, archaea that grow by oxidizing sulfur compounds release this element from rocks, making it available to other organisms. However, the archaea that do this, such as Sulfolobus, produce sulfuric acid as a waste product, and the growth of these organisms in abandoned mines can contribute to acid mine drainage and other environmental damage.[172]
In the carbon cycle, methanogen archaea remove hydrogen and play an important role in the decay of organic matter by the populations of microorganisms that act as decomposers in anaerobic ecosystems, such as sediments, marshes and sewage-treatment works.[173]
Global methane levels in 2011 had increased by a factor of 2.5 since pre-industrial times: from 722 ppb to 1800 ppb, the highest value in at least 800,000 years.[174] Methane has an anthropogenic global warming potential (AGWP) of 29, which means that it's 29 times stronger in heat-trapping than carbon dioxide is, over a 100-year time scale.[175]
Interactions with other organisms
The well-characterized interactions between archaea and other organisms are either mutual or commensal. There are no clear examples of known archaeal pathogens or parasites.[176][177] However, some species of methanogens have been suggested to be involved in infections in the mouth,[178][179] and Nanoarchaeum equitans may be a parasite of another species of archaea, since it only survives and reproduces within the cells of the Crenarchaeon Ignicoccus hospitalis,[180] and appears to offer no benefit to its host.[181] In contrast, Archaeal Richmond Mine Acidophilic Nanoorganisms (ARMAN) occasionally connect with other archaeal cells in acid mine drainage biofilms.[182] The nature of this relationship is unknown. However, it is distinct from that of Nanarchaeaum–Ignicoccus in that the ultrasmall ARMAN cells are usually seen independent of the Thermoplasmatales cells.
Mutualism
One well-understood example of mutualism is the interaction between protozoa and methanogenic archaea in the digestive tracts of animals that digest cellulose, such as ruminants and termites.[183] In these anaerobic environments, protozoa break down plant cellulose to obtain energy. This process releases hydrogen as a waste product, but high levels of hydrogen reduce energy production. When methanogens convert hydrogen to methane, protozoa benefit from more energy.[184]
In anaerobic protozoa, such as Plagiopyla frontata, archaea reside inside the protozoa and consume hydrogen produced in their hydrogenosomes.[185][186] Archaea also associate with larger organisms. For example, the marine archaean Cenarchaeum symbiosum lives within (is an endosymbiont of) the sponge Axinella mexicana.[187]
Commensalism
Archaea can also be commensals, benefiting from an association without helping or harming the other organism. For example, the methanogen Methanobrevibacter smithii is by far the most common archaean in the human flora, making up about one in ten of all the prokaryotes in the human gut.[188] In termites and in humans, these methanogens may in fact be mutualists, interacting with other microbes in the gut to aid digestion.[189] Archaean communities also associate with a range of other organisms, such as on the surface of corals,[190] and in the region of soil that surrounds plant roots (the rhizosphere).[191][192]
Significance in technology and industry
Extremophile archaea, particularly those resistant either to heat or to extremes of acidity and alkalinity, are a source of enzymes that function under these harsh conditions.[193][194] These enzymes have found many uses. For example, thermostable DNA polymerases, such as the Pfu DNA polymerase from Pyrococcus furiosus, revolutionized molecular biology by allowing the polymerase chain reaction to be used in research as a simple and rapid technique for cloning DNA. In industry, amylases, galactosidases and pullulanases in other species of Pyrococcus that function at over 100 °C (212 °F) allow food processing at high temperatures, such as the production of low lactose milk and whey.[195] Enzymes from these thermophilic archaea also tend to be very stable in organic solvents, allowing their use in environmentally friendly processes in green chemistry that synthesize organic compounds.[194] This stability makes them easier to use in structural biology. Consequently, the counterparts of bacterial or eukaryotic enzymes from extremophile archaea are often used in structural studies.[196]
In contrast to the range of applications of archaean enzymes, the use of the organisms themselves in biotechnology is less developed. Methanogenic archaea are a vital part of sewage treatment, since they are part of the community of microorganisms that carry out anaerobic digestion and produce biogas.[197] In mineral processing, acidophilic archaea display promise for the extraction of metals from ores, including gold, cobalt and copper.[198]
Archaea host a new class of potentially useful antibiotics. A few of these archaeocins have been characterized, but hundreds more are believed to exist, especially within Haloarchaea and Sulfolobus. These compounds differ in structure from bacterial antibiotics, so they may have novel modes of action. In addition, they may allow the creation of new selectable markers for use in archaeal molecular biology.[199]
See also
- Aerobic methane production
- List of Archaea genera
- List of sequenced archaeal genomes
- The Surprising Archaea (book)
- Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya
- Unique properties of hyperthermophilic archaea
References
- 1 2 3 Woese CR; Kandler O; Wheelis ML (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–9. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159
 . PMID 2112744.
. PMID 2112744. - ↑ Petitjean, C.; Deschamps, P.; López-García, P. & Moreira, D. (2014). "Rooting the Domain archaea by phylogenomic analysis supports the foundation of the new kingdom proteoarchaeota". Genome Biol. Evol. 7 (1): 191–204. doi:10.1093/gbe/evu274. PMC 4316627
 . PMID 25527841.
. PMID 25527841. - ↑ "NCBI taxonomy page on Archaea".
- ↑ Pace NR (May 2006). "Time for a change". Nature. 441 (7091): 289. Bibcode:2006Natur.441..289P. doi:10.1038/441289a. PMID 16710401.
- ↑ Stoeckenius W (1 October 1981). "Walsby's square bacterium: fine structure of an orthogonal procaryote". J. Bacteriol. 148 (1): 352–60. PMC 216199
 . PMID 7287626.
. PMID 7287626. - ↑ Bang C, Schmitz RA (2015). "Archaea associated with human surfaces: not to be underestimated". FEMS Microbiology Reviews. 39 (5): 631–48. doi:10.1093/femsre/fuv010. PMID 25907112.
- ↑ Staley JT (2006). "The bacterial species dilemma and the genomic-phylogenetic species concept". Philosophical Transactions of the Royal Society B. 361 (1475): 1899–909. doi:10.1098/rstb.2006.1914. PMC 1857736
 . PMID 17062409.
. PMID 17062409. - ↑ Zuckerkandl E; Pauling L (1965). "Molecules as documents of evolutionary history". J. Theor. Biol. 8 (2): 357–66. doi:10.1016/0022-5193(65)90083-4. PMID 5876245.
- ↑ Woese C; Fox G (1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 5088–90. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. PMC 432104
 . PMID 270744.
. PMID 270744. - ↑ Archaea. (2008). In Merriam-Webster Online Dictionary. Retrieved July 1, 2008
- ↑ Magrum LJ; Luehrsen KR; Woese CR (1978). "Are extreme halophiles actually "bacteria"?". J Mol Evol. 11 (1): 1–10. doi:10.1007/bf01768019. PMID 660662.
- ↑ Stetter KO. (1996). "Hyperthermophiles in the history of life.". Ciba Found Symp. 202: 1–10. PMID 9243007.
- 1 2 3 DeLong EF (1998). "Everything in moderation: archaea as 'non-extremophiles'". Current Opinion in Genetics & Development. 8 (6): 649–54. doi:10.1016/S0959-437X(98)80032-4. PMID 9914204.
- ↑ Theron J; Cloete TE (2000). "Molecular techniques for determining microbial diversity and community structure in natural environments". Crit. Rev. Microbiol. 26 (1): 37–57. doi:10.1080/10408410091154174. PMID 10782339.
- ↑ Schmidt TM (2006). "The maturing of microbial ecology" (PDF). Int. Microbiol. 9 (3): 217–23. PMID 17061212. Archived from the original (PDF) on 11 September 2008.
- ↑ Gevers D; Dawyndt P; Vandamme P; et al. (2006). "Stepping stones towards a new prokaryotic taxonomy". Philosophical Transactions of the Royal Society B. 361 (1475): 1911–6. doi:10.1098/rstb.2006.1915. PMC 1764938
 . PMID 17062410.
. PMID 17062410. - 1 2 Robertson CE; Harris JK; Spear JR; Pace NR (2005). "Phylogenetic diversity and ecology of environmental Archaea". Current Opinion in Microbiology. 8 (6): 638–42. doi:10.1016/j.mib.2005.10.003. PMID 16236543.
- ↑ Huber H; Hohn MJ; Rachel R; Fuchs T; Wimmer VC; Stetter KO (2002). "A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont". Nature. 417 (6884): 27–8. Bibcode:2002Natur.417...63H. doi:10.1038/417063a. PMID 11986665.
- ↑ Barns SM; Delwiche CF; Palmer JD; Pace NR (1996). "Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences". Proceedings of the National Academy of Sciences of the United States of America. 93 (17): 9188–93. Bibcode:1996PNAS...93.9188B. doi:10.1073/pnas.93.17.9188. PMC 38617
 . PMID 8799176.
. PMID 8799176. - ↑ Elkins JG; et al. (June 2008). "A korarchaeal genome reveals insights into the evolution of the Archaea". Proceedings of the National Academy of Sciences of the United States of America. 105 (23): 8102–7. Bibcode:2008PNAS..105.8102E. doi:10.1073/pnas.0801980105. PMC 2430366
 . PMID 18535141.
. PMID 18535141. - ↑ Baker, B.J.; Tyson, G.W.; Webb, R.I.; Flanagan, J.; Hugenholtz, P. & Banfield, J.F. (2006). "Lineages of acidophilic Archaea revealed by community genomic analysis. Science". Science. 314 (6884): 1933–1935. Bibcode:2006Sci...314.1933B. doi:10.1126/science.1132690. PMID 17185602.
- ↑ Baker BJ; et al. (May 2010). "Enigmatic, ultrasmall, uncultivated Archaea". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8806–11. Bibcode:2010PNAS..107.8806B. doi:10.1073/pnas.0914470107. PMC 2889320
 . PMID 20421484.
. PMID 20421484. - ↑ Guy, L; Ettema, TJ (19 December 2011). "The archaeal 'TACK' superphylum and the origin of eukaryotes.". Trends Microbiol. 19 (12): 580–587. doi:10.1016/j.tim.2011.09.002. PMID 22018741.
- ↑ de Queiroz K (2005). "Ernst Mayr and the modern concept of species". Proceedings of the National Academy of Sciences of the United States of America. 102 (Suppl 1): 6600–7. Bibcode:2005PNAS..102.6600D. doi:10.1073/pnas.0502030102. PMC 1131873
 . PMID 15851674.
. PMID 15851674. - ↑ Eppley JM; Tyson GW; Getz WM; Banfield JF (2007). "Genetic exchange across a species boundary in the archaeal genus ferroplasma". Genetics. 177 (1): 407–16. doi:10.1534/genetics.107.072892. PMC 2013692
 . PMID 17603112.
. PMID 17603112. - ↑ Papke RT; Zhaxybayeva O; Feil EJ; Sommerfeld K; Muise D; Doolittle WF (2007). "Searching for species in haloarchaea". Proceedings of the National Academy of Sciences of the United States of America. 104 (35): 14092–7. Bibcode:2007PNAS..10414092P. doi:10.1073/pnas.0706358104. PMC 1955782
 . PMID 17715057.
. PMID 17715057. - ↑ Kunin V; Goldovsky L; Darzentas N; Ouzounis CA (2005). "The net of life: reconstructing the microbial phylogenetic network". Genome Res. 15 (7): 954–9. doi:10.1101/gr.3666505. PMC 1172039
 . PMID 15965028.
. PMID 15965028. - ↑ Hugenholtz P (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biol. 3 (2): REVIEWS0003. doi:10.1186/gb-2002-3-2-reviews0003. PMC 139013
 . PMID 11864374.
. PMID 11864374. - ↑ Rappé MS; Giovannoni SJ (2003). "The uncultured microbial majority". Annu. Rev. Microbiol. 57: 369–94. doi:10.1146/annurev.micro.57.030502.090759. PMID 14527284.
- ↑ "Age of the Earth". U.S. Geological Survey. 1997. Archived from the original on 23 December 2005. Retrieved 2006-01-10.
- ↑ Dalrymple, G. Brent (2001). "The age of the Earth in the twentieth century: a problem (mostly) solved". Special Publications, Geological Society of London. 190 (1): 205–221. Bibcode:2001GSLSP.190..205D. doi:10.1144/GSL.SP.2001.190.01.14.
- ↑ Manhesa, Gérard; Allègre, Claude J.; Dupréa, Bernard & Hamelin, Bruno (1980). "Lead isotope study of basic-ultrabasic layered complexes: Speculations about the age of the earth and primitive mantle characteristics". Earth and Planetary Science Letters. 47 (3): 370–382. Bibcode:1980E&PSL..47..370M. doi:10.1016/0012-821X(80)90024-2.
- ↑ de Duve, Christian (October 1995). "The Beginnings of Life on Earth". American Scientist. Retrieved 15 January 2014.
- ↑ Timmer, John (4 September 2012). "3.5 billion year old organic deposits show signs of life". Ars Technica. Retrieved 15 January 2014.
- ↑ Yoko Ohtomo; Takeshi Kakegawa; Akizumi Ishida; Toshiro Nagase; Minik T. Rosing (8 December 2013). "Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks". Nature Geoscience. Nature Geoscience. 7: 25. Bibcode:2014NatGe...7...25O. doi:10.1038/ngeo2025. Retrieved 9 Dec 2013.
- ↑ Borenstein, Seth (13 November 2013). "Oldest fossil found: Meet your microbial mom". Associated Press. Retrieved 15 November 2013.
- ↑ Noffke, Nora; Christian, Daniel; Wacey, David; Hazen, Robert M. (8 November 2013). "Microbially Induced Sedimentary Structures Recording an Ancient Ecosystem in the ca. 3.48 Billion-Year-Old Dresser Formation, Pilbara, Western Australia". Astrobiology (journal). 13 (12): 1103–24. Bibcode:2013AsBio..13.1103N. doi:10.1089/ast.2013.1030. PMC 3870916
 . PMID 24205812. Retrieved 15 November 2013.
. PMID 24205812. Retrieved 15 November 2013. - 1 2 Borenstein, Seth (19 October 2015). "Hints of life on what was thought to be desolate early Earth". Excite. Yonkers, NY: Mindspark Interactive Network. Associated Press. Retrieved 2015-10-20.
- ↑ Bell, Elizabeth A.; Boehnike, Patrick; Harrison, T. Mark; et al. (19 October 2015). "Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon" (PDF). Proc. Natl. Acad. Sci. U.S.A. Washington, D.C.: National Academy of Sciences. 112 (47): 14518–21. Bibcode:2015PNAS..11214518B. doi:10.1073/pnas.1517557112. ISSN 1091-6490. PMC 4664351
 . PMID 26483481. Retrieved 2015-10-20. Early edition, published online before print.
. PMID 26483481. Retrieved 2015-10-20. Early edition, published online before print. - ↑ Schopf J (2006). "Fossil evidence of Archaean life" (PDF). Philosophical Transactions of the Royal Society B. 361 (1470): 869–85. doi:10.1098/rstb.2006.1834. PMC 1578735
 . PMID 16754604.
. PMID 16754604. - ↑ Chappe B; Albrecht P; Michaelis W (July 1982). "Polar Lipids of Archaebacteria in Sediments and Petroleums". Science. 217 (4554): 65–66. Bibcode:1982Sci...217...65C. doi:10.1126/science.217.4554.65. PMID 17739984.
- ↑ Brocks JJ; Logan GA; Buick R; Summons RE (1999). "Archean molecular fossils and the early rise of eukaryotes". Science. 285 (5430): 1033–6. doi:10.1126/science.285.5430.1033. PMID 10446042.
- ↑ Rasmussen B; Fletcher IR; Brocks JJ; Kilburn MR (October 2008). "Reassessing the first appearance of eukaryotes and cyanobacteria". Nature. 455 (7216): 1101–4. Bibcode:2008Natur.455.1101R. doi:10.1038/nature07381. PMID 18948954.
- ↑ Hahn, Jürgen; Pat Haug (1986). "Traces of Archaebacteria in ancient sediments". System Applied Microbiology. 7 (Archaebacteria '85 Proceedings): 178–83. doi:10.1016/S0723-2020(86)80002-9.
- ↑ Wang M; Yafremava LS; Caetano-Anollés D; Mittenthal JE; Caetano-Anollés G (2007). "Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world". Genome Res. 17 (11): 1572–85. doi:10.1101/gr.6454307. PMC 2045140
 . PMID 17908824.
. PMID 17908824. - ↑ Woese CR; Gupta R (1981). "Are archaebacteria merely derived 'prokaryotes'?". Nature. 289 (5793): 95–6. Bibcode:1981Natur.289...95W. doi:10.1038/289095a0. PMID 6161309.
- 1 2 3 Woese C (1998). "The universal ancestor". Proceedings of the National Academy of Sciences of the United States of America. 95 (12): 6854–9. Bibcode:1998PNAS...95.6854W. doi:10.1073/pnas.95.12.6854. PMC 22660
 . PMID 9618502.
. PMID 9618502. - 1 2 Kandler O. The early diversification of life and the origin of the three domains: A proposal. In: Wiegel J, Adams WW, editors. Thermophiles: The keys to molecular evolution and the origin of life? Athens: Taylor and Francis, 1998: 19-31.
- ↑ Gribaldo S; Brochier-Armanet C (2006). "The origin and evolution of Archaea: a state of the art". Philosophical Transactions of the Royal Society B. 361 (1470): 1007–22. doi:10.1098/rstb.2006.1841. PMC 1578729
 . PMID 16754611.
. PMID 16754611. - 1 2 Woese CR (1 March 1994). "There must be a prokaryote somewhere: microbiology's search for itself". Microbiol. Rev. 58 (1): 1–9. PMC 372949
 . PMID 8177167.
. PMID 8177167. - ↑ Information is from Willey JM, Sherwood LM, Woolverton CJ. Microbiology 7th ed. (2008), Ch. 19 pp. 474–475, except where noted.
- ↑ Jurtshuk, Peter (1996). Medical Microbiology (4th ed.). Galveston (TX): University of Texas Medical Branch at Galveston. Retrieved 5 November 2014.
- 1 2 3 Woese C; Fox G (1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 5088–90. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. PMC 432104
 . PMID 270744.
. PMID 270744. - ↑ Howland, John L. (2000). The Surprising Archaea: Discovering Another Domain of Life. Oxford: Oxford University Press. pp. 25–30. ISBN 0-19-511183-4.
- 1 2 3 Cavicchioli, Ricardo (January 2011). "Archaea- timeline of the third domain". Nature Reviews Microbiology. 9 (1): 51–61. doi:10.1038/nrmicro2482. PMID 21132019. Retrieved 5 November 2014.
- ↑ Deppenmeier, U. (2002). "The unique biochemistry of methanogenesis". Prog Nucleic Acid Res Mol Biol. Progress in Nucleic Acid Research and Molecular Biology. 71: 223–283. doi:10.1016/s0079-6603(02)71045-3. ISBN 9780125400718. PMID 12102556.
- ↑ Koonin EV; Mushegian AR; Galperin MY; Walker DR (1997). "Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea". Mol Microbiol. 25 (4): 619–637. doi:10.1046/j.1365-2958.1997.4821861.x. PMID 9379893.
- 1 2 3 4 5 Gupta R. S. (1998). "Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes". Microbiol. Mol. Biol. Rev. 62 (4): 1435–1491. PMC 98952
 . PMID 9841678.
. PMID 9841678. - ↑ Koch AL (2003). "Were Gram-positive rods the first bacteria?". Trends Microbiol. 11 (4): 166–170. doi:10.1016/S0966-842X(03)00063-5. PMID 12706994.
- 1 2 3 Gupta R.S. (1998). "What are archaebacteria: life's third domain or monoderm prokaryotes related to gram-positive bacteria? A new proposal for the classification of prokaryotic organisms". Mol. Microbiol. 29 (3): 695–708. doi:10.1046/j.1365-2958.1998.00978.x. PMID 9723910.
- ↑ Gogarten, Johann Peter (1994). "Which is the Most Conserved Group of Proteins? Homology - Orthology, Paralogy, Xenology and the Fusion of Independent Lineages.". Journal of Molecular Evolution. 39 (5): 541–543. doi:10.1007/bf00173425. PMID 7807544.
- ↑ Brown JR; Masuchi Y; Robb FT; Doolittle WF (1994). "Evolutionary relationships of bacterial and archaeal glutamine synthetase genes". J Mol Evol. 38 (6): 566–576. doi:10.1007/BF00175876. PMID 7916055.
- 1 2 3 Gupta R.S. (2000). "The natural evolutionary relationships among prokaryotes". Crit. Rev. Microbiol. 26 (2): 111–131. doi:10.1080/10408410091154219. PMID 10890353.
- ↑ Gupta RS. Molecular Sequences and the Early History of Life. In: Sapp J, editor. Microbial Phylogeny and Evolution: Concepts and Controversies. New York: Oxford University Press, 2005: 160-183.
- ↑ Cavalier-Smith T (2002). "The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification". Int J Syst Evol Microbiol. 52 (1): 7–76. doi:10.1099/00207713-52-1-7. PMID 11837318.
- ↑ Valas RE; Bourne PE (2011). "The origin of a derived superkingdom: how a Gram-positive bacterium crossed the desert to become an archaeon". Biol Direct. 6: 16. doi:10.1186/1745-6150-6-16. PMC 3056875
 . PMID 21356104.
. PMID 21356104. - ↑ Skophammer RG; Herbold CW; Rivera MC; Servin JA; Lake JA (2006). "Evidence that the root of the tree of life is not within the Archaea". Mol Biol Evol. 23 (9): 1648–1651. doi:10.1093/molbev/msl046. PMID 16801395.
- ↑ Lake JA (January 1988). "Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences". Nature. 331 (6152): 184–6. Bibcode:1988Natur.331..184L. doi:10.1038/331184a0. PMID 3340165.
- ↑ Nelson KE; et al. (1999). "Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima". Nature. 399 (6734): 323–9. Bibcode:1999Natur.399..323N. doi:10.1038/20601. PMID 10360571.
- ↑ Gouy M; Li WH (May 1989). "Phylogenetic analysis based on rRNA sequences supports the archaebacterial rather than the eocyte tree". Nature. 339 (6220): 145–7. Bibcode:1989Natur.339..145G. doi:10.1038/339145a0. PMID 2497353.
- ↑ Yutin N; Makarova KS; Mekhedov SL; Wolf YI; Koonin EV (May 2008). "The deep archaeal roots of eukaryotes". Mol. Biol. Evol. 25 (8): 1619–30. doi:10.1093/molbev/msn108. PMC 2464739
 . PMID 18463089.
. PMID 18463089. - ↑ Lake JA. (1988). "Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences". Nature. 331 (6152): 184–6. Bibcode:1988Natur.331..184L. doi:10.1038/331184a0. PMID 3340165.
- ↑ Williams, Tom A.; Foster, Peter G.; Cox, Cymon J.; Embley, T. Martin (December 2013). "An archaeal origin of eukaryotes supports only two primary domains of life". Nature. 504 (7479): 231–236. Bibcode:2013Natur.504..231W. doi:10.1038/nature12779. PMID 24336283.
- ↑ Zimmer, Carl (May 6, 2015). "Under the Sea, a Missing Link in the Evolution of Complex Cells". New York Times. Retrieved May 6, 2015.
- ↑ Spang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; Eijk, Roel van; Schleper, Christa; Guy, Lionel (2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature. 521 (7551): 173–179. Bibcode:2015Natur.521..173S. doi:10.1038/nature14447. PMC 4444528
 . PMID 25945739.
. PMID 25945739. - 1 2 3 4 Krieg, Noel (2005). Bergey's Manual of Systematic Bacteriology. US: Springer. pp. 21–6. ISBN 978-0-387-24143-2.
- ↑ Barns, Sue and Burggraf, Siegfried. (1997) Crenarchaeota. Version 1 January 1997. in The Tree of Life Web Project
- ↑ Walsby, A.E. (1980). "A square bacterium". Nature. 283 (5742): 69–71. Bibcode:1980Natur.283...69W. doi:10.1038/283069a0.
- ↑ Hara F; Yamashiro K; Nemoto N; et al. (2007). "An actin homolog of the archaeon Thermoplasma acidophilum that retains the ancient characteristics of eukaryotic actin". J. Bacteriol. 189 (5): 2039–45. doi:10.1128/JB.01454-06. PMC 1855749
 . PMID 17189356.
. PMID 17189356. - ↑ Trent JD; Kagawa HK; Yaoi T; Olle E; Zaluzec NJ (1997). "Chaperonin filaments: the archaeal cytoskeleton?". Proceedings of the National Academy of Sciences of the United States of America. 94 (10): 5383–8. Bibcode:1997PNAS...94.5383T. doi:10.1073/pnas.94.10.5383. PMC 24687
 . PMID 9144246.
. PMID 9144246. - ↑ Hixon WG; Searcy DG (1993). "Cytoskeleton in the archaebacterium Thermoplasma acidophilum? Viscosity increase in soluble extracts". BioSystems. 29 (2–3): 151–60. doi:10.1016/0303-2647(93)90091-P. PMID 8374067.
- 1 2 Golyshina OV; Pivovarova TA; Karavaiko GI; et al. (1 May 2000). "Ferroplasma acidiphilum gen. nov., sp. nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea". Int. J. Syst. Evol. Microbiol. 50 (3): 997–1006. doi:10.1099/00207713-50-3-997. PMID 10843038.
- ↑ Hall-Stoodley L; Costerton JW; Stoodley P (2004). "Bacterial biofilms: from the natural environment to infectious diseases". Nature Reviews Microbiology. 2 (2): 95–108. doi:10.1038/nrmicro821. PMID 15040259.
- ↑ Kuwabara T; Minaba M; Iwayama Y; et al. (November 2005). "Thermococcus coalescens sp. nov., a cell-fusing hyperthermophilic archaeon from Suiyo Seamount". Int. J. Syst. Evol. Microbiol. 55 (Pt 6): 2507–14. doi:10.1099/ijs.0.63432-0. PMID 16280518.
- ↑ Nickell S; Hegerl R; Baumeister W; Rachel R (2003). "Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography". J. Struct. Biol. 141 (1): 34–42. doi:10.1016/S1047-8477(02)00581-6. PMID 12576018.
- ↑ Horn C; Paulmann B; Kerlen G; Junker N; Huber H (15 August 1999). "In vivo observation of cell division of anaerobic hyperthermophiles by using a high-intensity dark-field microscope". J. Bacteriol. 181 (16): 5114–8. PMC 94007
 . PMID 10438790.
. PMID 10438790. - ↑ Rudolph C; Wanner G; Huber R (May 2001). "Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology". Appl. Environ. Microbiol. 67 (5): 2336–44. doi:10.1128/AEM.67.5.2336-2344.2001. PMC 92875
 . PMID 11319120.
. PMID 11319120. - 1 2 Thomas NA; Bardy SL; Jarrell KF (2001). "The archaeal flagellum: a different kind of prokaryotic motility structure". FEMS Microbiol. Rev. 25 (2): 147–74. doi:10.1111/j.1574-6976.2001.tb00575.x. PMID 11250034.
- ↑ Rachel R; Wyschkony I; Riehl S; Huber H (March 2002). "The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon". Archaea. 1 (1): 9–18. doi:10.1155/2002/307480. PMC 2685547
 . PMID 15803654.
. PMID 15803654. - ↑ Sára M; Sleytr UB (2000). "S-Layer proteins". J. Bacteriol. 182 (4): 859–68. doi:10.1128/JB.182.4.859-868.2000. PMC 94357
 . PMID 10648507.
. PMID 10648507. - ↑ Engelhardt H; Peters J (1998). "Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions". J Struct Biol. 124 (2–3): 276–302. doi:10.1006/jsbi.1998.4070. PMID 10049812.
- 1 2 Kandler, O; König, H (1998). "Cell wall polymers in Archaea (Archaebacteria)" (PDF). Cellular and Molecular Life Sciences (CMLS). 54 (4): 305–308. doi:10.1007/s000180050156.
- ↑ Howland, John L. (2000). The Surprising Archaea: Discovering Another Domain of Life. Oxford: Oxford University Press. p. 32. ISBN 0-19-511183-4.
- ↑ Gophna U; Ron EZ; Graur D (July 2003). "Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events". Gene. 312: 151–63. doi:10.1016/S0378-1119(03)00612-7. PMID 12909351.
- ↑ Nguyen L; Paulsen IT; Tchieu J; Hueck CJ; Saier MH (April 2000). "Phylogenetic analyses of the constituents of Type III protein secretion systems". J. Mol. Microbiol. Biotechnol. 2 (2): 125–44. PMID 10939240.
- ↑ Ng SY; Chaban B; Jarrell KF (2006). "Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications". J. Mol. Microbiol. Biotechnol. 11 (3–5): 167–91. doi:10.1159/000094053. PMID 16983194.
- ↑ Bardy SL; Ng SY; Jarrell KF (February 2003). "Prokaryotic motility structures". Microbiology (Reading, Engl.). 149 (Pt 2): 295–304. doi:10.1099/mic.0.25948-0. PMID 12624192.
- 1 2 Koga Y; Morii H (2007). "Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations". Microbiol. Mol. Biol. Rev. 71 (1): 97–120. doi:10.1128/MMBR.00033-06. PMC 1847378
 . PMID 17347520.
. PMID 17347520. - ↑ De Rosa M; Gambacorta A; Gliozzi A (1 March 1986). "Structure, biosynthesis, and physicochemical properties of archaebacterial lipids". Microbiol. Rev. 50 (1): 70–80. PMC 373054
 . PMID 3083222.
. PMID 3083222. - ↑ Albers SV; van de Vossenberg JL; Driessen AJ; Konings WN (September 2000). "Adaptations of the archaeal cell membrane to heat stress". Front. Biosci. 5: D813–20. doi:10.2741/albers. PMID 10966867.
- ↑ Damsté JS; Schouten S; Hopmans EC; van Duin AC; Geenevasen JA (October 2002). "Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota". J. Lipid Res. 43 (10): 1641–51. doi:10.1194/jlr.M200148-JLR200. PMID 12364548.
- ↑ Koga Y; Morii H (November 2005). "Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects". Biosci. Biotechnol. Biochem. 69 (11): 2019–34. doi:10.1271/bbb.69.2019. PMID 16306681.
- ↑ Hanford MJ; Peeples TL (January 2002). "Archaeal tetraether lipids: unique structures and applications". Appl. Biochem. Biotechnol. 97 (1): 45–62. doi:10.1385/ABAB:97:1:45. PMID 11900115.
- ↑ Macalady JL; Vestling MM; Baumler D; Boekelheide N; Kaspar CW; Banfield JF (October 2004). "Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid". Extremophiles. 8 (5): 411–9. doi:10.1007/s00792-004-0404-5. PMID 15258835.
- 1 2 3 Valentine DL (2007). "Adaptations to energy stress dictate the ecology and evolution of the Archaea". Nature Reviews Microbiology. 5 (4): 316–23. doi:10.1038/nrmicro1619. PMID 17334387.
- 1 2 3 Schäfer G; Engelhard M; Müller V (1 September 1999). "Bioenergetics of the Archaea". Microbiol. Mol. Biol. Rev. 63 (3): 570–620. PMC 103747
 . PMID 10477309.
. PMID 10477309. - ↑ Zillig W (December 1991). "Comparative biochemistry of Archaea and Bacteria". Current Opinion in Genetics & Development. 1 (4): 544–51. doi:10.1016/S0959-437X(05)80206-0. PMID 1822288.
- ↑ Romano A; Conway T (1996). "Evolution of carbohydrate metabolic pathways". Res Microbiol. 147 (6–7): 448–55. doi:10.1016/0923-2508(96)83998-2. PMID 9084754.
- ↑ Koch A (1998). "How did bacteria come to be?". Adv Microb Physiol. Advances in Microbial Physiology. 40: 353–99. doi:10.1016/S0065-2911(08)60135-6. ISBN 978-0-12-027740-7. PMID 9889982.
- ↑ DiMarco AA; Bobik TA; Wolfe RS (1990). "Unusual coenzymes of methanogenesis". Annu. Rev. Biochem. 59: 355–94. doi:10.1146/annurev.bi.59.070190.002035. PMID 2115763.
- ↑ Klocke M; Nettmann E; Bergmann I; et al. (May 2008). "Characterization of the methanogenic Archaea within two-phase biogas reactor systems operated with plant biomass". Syst. Appl. Microbiol. 31 (3): 190–205. doi:10.1016/j.syapm.2008.02.003. PMID 18501543.
- ↑ Based on PDB 1FBB. Data published in Subramaniam S; Henderson R (August 2000). "Molecular mechanism of vectorial proton translocation by bacteriorhodopsin". Nature. 406 (6796): 653–7. doi:10.1038/35020614. PMID 10949309.
- ↑ Mueller-Cajar O; Badger MR (August 2007). "New roads lead to Rubisco in archaebacteria". BioEssays. 29 (8): 722–4. doi:10.1002/bies.20616. PMID 17621634.
- ↑ Berg IA; Kockelkorn D; Buckel W; Fuchs G (December 2007). "A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea". Science. 318 (5857): 1782–6. Bibcode:2007Sci...318.1782B. doi:10.1126/science.1149976. PMID 18079405.
- ↑ Thauer RK (December 2007). "Microbiology. A fifth pathway of carbon fixation". Science. 318 (5857): 1732–3. doi:10.1126/science.1152209. PMID 18079388.
- ↑ Bryant DA; Frigaard NU (November 2006). "Prokaryotic photosynthesis and phototrophy illuminated". Trends Microbiol. 14 (11): 488–96. doi:10.1016/j.tim.2006.09.001. PMID 16997562.
- ↑ Könneke M; Bernhard AE; de la Torre JR; Walker CB; Waterbury JB; Stahl DA (September 2005). "Isolation of an autotrophic ammonia-oxidizing marine archaeon". Nature. 437 (7058): 543–6. Bibcode:2005Natur.437..543K. doi:10.1038/nature03911. PMID 16177789.
- 1 2 Francis CA; Beman JM; Kuypers MM (May 2007). "New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation". ISME J. 1 (1): 19–27. doi:10.1038/ismej.2007.8. PMID 18043610.
- ↑ Lanyi JK (2004). "Bacteriorhodopsin". Annu. Rev. Physiol. 66: 665–88. doi:10.1146/annurev.physiol.66.032102.150049. PMID 14977418.
- 1 2 Allers T; Mevarech M (2005). "Archaeal genetics — the third way". Nature Reviews Genetics. 6 (1): 58–73. doi:10.1038/nrg1504. PMID 15630422.
- ↑ Galagan JE; Nusbaum C; Roy A; et al. (April 2002). "The genome of M. acetivorans reveals extensive metabolic and physiological diversity". Genome Res. 12 (4): 532–42. doi:10.1101/gr.223902. PMC 187521
 . PMID 11932238.
. PMID 11932238. - ↑ Waters E; et al. (2003). "The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism". Proceedings of the National Academy of Sciences of the United States of America. 100 (22): 12984–8. Bibcode:2003PNAS..10012984W. doi:10.1073/pnas.1735403100. PMC 240731
 . PMID 14566062.
. PMID 14566062. - ↑ Schleper C; Holz I; Janekovic D; Murphy J; Zillig W (1 August 1995). "A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating". J. Bacteriol. 177 (15): 4417–26. PMC 177192
 . PMID 7635827.
. PMID 7635827. - ↑ Sota M; Top EM (2008). "Horizontal Gene Transfer Mediated by Plasmids". Plasmids: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-35-6.
- ↑ Xiang X; Chen L; Huang X; Luo Y; She Q; Huang L (2005). "Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features". J. Virol. 79 (14): 8677–86. doi:10.1128/JVI.79.14.8677-8686.2005. PMC 1168784
 . PMID 15994761.
. PMID 15994761. - ↑ Prangishvili D; Forterre P; Garrett RA (2006). "Viruses of the Archaea: a unifying view". Nature Reviews Microbiology. 4 (11): 837–48. doi:10.1038/nrmicro1527. PMID 17041631.
- ↑ Prangishvili D; Garrett RA (2004). "Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses". Biochem. Soc. Trans. 32 (Pt 2): 204–8. doi:10.1042/BST0320204. PMID 15046572.
- ↑ Pietilä MK; Roine E; Paulin L; Kalkkinen N; Bamford DH (March 2009). "An ssDNA virus infecting archaea; A new lineage of viruses with a membrane envelope". Mol. Microbiol. 72 (2): 307–19. doi:10.1111/j.1365-2958.2009.06642.x. PMID 19298373.
- ↑ Mochizuki T; Krupovic M; Pehau-Arnaudet G; Sako Y; Forterre P; Prangishvili D (2012). "Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome". Proceedings of the National Academy of Sciences of the United States of America. 109 (33): 13386–13391. Bibcode:2012PNAS..10913386M. doi:10.1073/pnas.1203668109. PMC 3421227
 . PMID 22826255.
. PMID 22826255. - ↑ Mojica FJ; Díez-Villaseñor C; García-Martínez J; Soria E (2005). "Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements". J. Mol. Evol. 60 (2): 174–82. doi:10.1007/s00239-004-0046-3. PMID 15791728.
- ↑ Makarova KS; Grishin NV; Shabalina SA; Wolf YI; Koonin EV (2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biol. Direct. 1: 7. doi:10.1186/1745-6150-1-7. PMC 1462988
 . PMID 16545108.
. PMID 16545108. - ↑ Graham DE; Overbeek R; Olsen GJ; Woese CR (2000). "An archaeal genomic signature". Proceedings of the National Academy of Sciences of the United States of America. 97 (7): 3304–8. Bibcode:2000PNAS...97.3304G. doi:10.1073/pnas.050564797. PMC 16234
 . PMID 10716711.
. PMID 10716711. - 1 2 Gaasterland T (1999). "Archaeal genomics". Current Opinion in Microbiology. 2 (5): 542–7. doi:10.1016/S1369-5274(99)00014-4. PMID 10508726.
- ↑ Dennis PP (1997). "Ancient Ciphers: Translation in Archaea". Cell. 89 (7): 1007–10. doi:10.1016/S0092-8674(00)80288-3. PMID 9215623.
- ↑ Werner F (September 2007). "Structure and function of archaeal RNA polymerases". Mol. Microbiol. 65 (6): 1395–404. doi:10.1111/j.1365-2958.2007.05876.x. PMID 17697097.
- ↑ Aravind L; Koonin EV (1999). "DNA-binding proteins and evolution of transcription regulation in the archaea". Nucleic Acids Res. 27 (23): 4658–70. doi:10.1093/nar/27.23.4658. PMC 148756
 . PMID 10556324.
. PMID 10556324. - ↑ Lykke-Andersen J; Aagaard C; Semionenkov M; Garrett RA (September 1997). "Archaeal introns: splicing, intercellular mobility and evolution". Trends Biochem. Sci. 22 (9): 326–31. doi:10.1016/S0968-0004(97)01113-4. PMID 9301331.
- ↑ Watanabe Y; Yokobori S; Inaba T; et al. (January 2002). "Introns in protein-coding genes in Archaea". FEBS Lett. 510 (1–2): 27–30. doi:10.1016/S0014-5793(01)03219-7. PMID 11755525.
- ↑ Yoshinari S; Itoh T; Hallam SJ; et al. (August 2006). "Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease". Biochem. Biophys. Res. Commun. 346 (3): 1024–32. doi:10.1016/j.bbrc.2006.06.011. PMID 16781672.
- ↑ Rosenshine, I; Tchelet, R; Mevarech, M. (1989). "The mechanism of DNA transfer in the mating system of an archaebacterium". Science. 245 (4924): 1387–1389. Bibcode:1989Sci...245.1387R. doi:10.1126/science.2818746. PMID 2818746.
- 1 2 3 Fröls, S; Ajon, M; Wagner, M; Teichmann, D; Zolghadr, B; Folea, M; Boekema, EJ; Driessen, AJ; Schleper, C; et al. (2008). "UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation". Mol Microbiol. 70 (4): 938–52. doi:10.1111/j.1365-2958.2008.06459.x. PMID 18990182.
- 1 2 3 Ajon, M; Fröls, S; van Wolferen, M; Stoecker, K; Teichmann, D; Driessen, AJ; Grogan, DW; Albers, SV; Schleper, C.; et al. (2011). "UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili". Mol Microbiol. 82 (4): 807–17. doi:10.1111/j.1365-2958.2011.07861.x. PMID 21999488.
- ↑ Fröls, S; White, MF; Schleper, C. (2009). "Reactions to UV damage in the model archaeon Sulfolobus solfataricus". Biochem Soc Trans. 37 (1): 36–41. doi:10.1042/BST0370036. PMID 19143598.
- ↑ Bernstein, H; Bernstein, C (2013). Bernstein, Carol, ed. Evolutionary Origin and Adaptive Function of Meiosis, Meiosis. InTech. ISBN 978-953-51-1197-9.
- 1 2 Bernander R (1998). "Archaea and the cell cycle". Mol. Microbiol. 29 (4): 955–61. doi:10.1046/j.1365-2958.1998.00956.x. PMID 9767564.
- ↑ Kelman LM; Kelman Z (2004). "Multiple origins of replication in archaea". Trends Microbiol. 12 (9): 399–401. doi:10.1016/j.tim.2004.07.001. PMID 15337158.
- ↑ Lindås AC; Karlsson EA; Lindgren MT; Ettema TJ; Bernander R. (2008). "A unique cell division machinery in the Archaea". Proc Natl Acad Sci U S A. 105 (48): 18942–6. Bibcode:2008PNAS..10518942L. doi:10.1073/pnas.0809467105. PMC 2596248
 . PMID 18987308.
. PMID 18987308. - ↑ Samson RY; Obita T; Freund SM; Williams RL; Bell SD. (2008). "A role for the ESCRT system in cell division in archaea". Science. 12 (322): 1710–3. Bibcode:2008Sci...322.1710S. doi:10.1126/science.1165322. PMC 4121953
 . PMID 19008417.
. PMID 19008417. - ↑ Pelve EA; Lindås AC; Martens-Habbena W; de la Torre JR; Stahl DA; Bernander R (2011). "Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus.". Mol Microbiol. 82 (3): 555–566. doi:10.1111/j.1365-2958.2011.07834.x. PMID 21923770.
- ↑ Onyenwoke RU; Brill JA; Farahi K; Wiegel J (2004). "Sporulation genes in members of the low G+C Gram-type-positive phylogenetic branch ( Firmicutes)". Arch. Microbiol. 182 (2–3): 182–92. doi:10.1007/s00203-004-0696-y. PMID 15340788.
- ↑ Kostrikina NA; Zvyagintseva IS; Duda VI (1991). "Cytological peculiarities of some extremely halophilic soil archaeobacteria". Arch. Microbiol. 156 (5): 344–49. doi:10.1007/BF00248708.
- ↑ DeLong EF; Pace NR (2001). "Environmental diversity of bacteria and archaea". Syst. Biol. 50 (4): 470–8. doi:10.1080/106351501750435040. PMID 12116647.
- 1 2 Pikuta EV; Hoover RB; Tang J (2007). "Microbial extremophiles at the limits of life". Crit. Rev. Microbiol. 33 (3): 183–209. doi:10.1080/10408410701451948. PMID 17653987.
- ↑ Madigan MT; Martino JM (2006). Brock Biology of Microorganisms (11th ed.). Pearson. p. 136. ISBN 0-13-196893-9.
- ↑ Takai K; Nakamura K; Toki T; Tsunogai U; Miyazaki M; Miyazaki J; Hirayama H; Nakagawa S; Nunoura T; Horikoshi K (2008). "Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation". Proceedings of the National Academy of Sciences of the United States of America. 105 (31): 10949–54. Bibcode:2008PNAS..10510949T. doi:10.1073/pnas.0712334105. PMC 2490668
 . PMID 18664583.
. PMID 18664583. - ↑ Ciaramella M; Napoli A; Rossi M (February 2005). "Another extreme genome: how to live at pH 0". Trends Microbiol. 13 (2): 49–51. doi:10.1016/j.tim.2004.12.001. PMID 15680761.
- ↑ Javaux EJ (2006). "Extreme life on Earth—past, present and possibly beyond". Res. Microbiol. 157 (1): 37–48. doi:10.1016/j.resmic.2005.07.008. PMID 16376523.
- ↑ Nealson KH (January 1999). "Post-Viking microbiology: new approaches, new data, new insights" (PDF). Origins of Life and Evolution of Biospheres. 29 (1): 73–93. doi:10.1023/A:1006515817767. PMID 11536899.
- ↑ Davies PC (1996). "The transfer of viable microorganisms between planets". Ciba Found. Symp. 202: 304–14; discussion 314–7. PMID 9243022.
- ↑ López-García P; López-López A; Moreira D; Rodríguez-Valera F (July 2001). "Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front". FEMS Microbiol. Ecol. 36 (2–3): 193–202. doi:10.1016/s0168-6496(01)00133-7. PMID 11451524.
- ↑ Karner MB; DeLong EF; Karl DM (2001). "Archaeal dominance in the mesopelagic zone of the Pacific Ocean". Nature. 409 (6819): 507–10. Bibcode:2001Natur.409..507K. doi:10.1038/35054051. PMID 11206545.
- ↑ Giovannoni SJ; Stingl U (2005). "Molecular diversity and ecology of microbial plankton". Nature. 427 (7057): 343–8. Bibcode:2005Natur.437..343G. doi:10.1038/nature04158. PMID 16163344.
- ↑ DeLong EF; Karl DM (September 2005). "Genomic perspectives in microbial oceanography". Nature. 437 (7057): 336–42. Bibcode:2005Natur.437..336D. doi:10.1038/nature04157. PMID 16163343.
- ↑ Konneke M; Bernhard AE; de la Torre JR; Walker CB; Waterbury JB; Stahl DA (2005). "Isolation of an autotrophic ammonia-oxidizing marine archaeon". Nature. 437 (7057): 543–6. Bibcode:2005Natur.437..543K. doi:10.1038/nature03911. PMID 16177789.
- ↑ Agogué, H; Brink, M; Dinasquet, J; Herndl, GJ (2008). "Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic". Nature. 456 (7223): 788–791. Bibcode:2008Natur.456..788A. doi:10.1038/nature07535. PMID 19037244.
- ↑ Teske A; Sørensen KB (January 2008). "Uncultured archaea in deep marine subsurface sediments: have we caught them all?". ISME J. 2 (1): 3–18. doi:10.1038/ismej.2007.90. PMID 18180743.
- ↑ Lipp JS; Morono Y; Inagaki F; Hinrichs KU (July 2008). "Significant contribution of Archaea to extant biomass in marine subsurface sediments". Nature. 454 (7207): 991–4. Bibcode:2008Natur.454..991L. doi:10.1038/nature07174. PMID 18641632.
- ↑ Cabello P; Roldán MD; Moreno-Vivián C (November 2004). "Nitrate reduction and the nitrogen cycle in archaea". Microbiology (Reading, Engl.). 150 (Pt 11): 3527–46. doi:10.1099/mic.0.27303-0. PMID 15528644.
- ↑ Mehta MP; Baross JA (December 2006). "Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon". Science. 314 (5806): 1783–6. Bibcode:2006Sci...314.1783M. doi:10.1126/science.1134772. PMID 17170307.
- ↑ Coolen MJ; Abbas B; van Bleijswijk J; et al. (April 2007). "Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids". Environ. Microbiol. 9 (4): 1001–16. doi:10.1111/j.1462-2920.2006.01227.x. PMID 17359272.
- ↑ Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G. W.; Prosser, J. I.; Schuster, S. C.; Schleper, C. (2006). "Archaea predominate among ammonia-oxidizing prokaryotes in soils". Nature. 442 (7104): 806–809. Bibcode:2006Natur.442..806L. doi:10.1038/nature04983. PMID 16915287.
- ↑ Baker, B. J; Banfield, J. F (2003). "Microbial communities in acid mine drainage". FEMS Microbiology Ecology. 44 (2): 139–152. doi:10.1016/S0168-6496(03)00028-X. PMID 19719632.
- ↑ Schimel J (August 2004). "Playing scales in the methane cycle: from microbial ecology to the globe". Proceedings of the National Academy of Sciences of the United States of America. 101 (34): 12400–1. Bibcode:2004PNAS..10112400S. doi:10.1073/pnas.0405075101. PMC 515073
 . PMID 15314221.
. PMID 15314221.
- ↑ Ipcc ar5 wg1 (2013). "Climate Change 2013: The Physical Science Basis - Summary for Policymakers" (PDF). Cambridge University Press.
- ↑ Ipcc ar5 wg1 (2013). "Climate Change 2013: The Physical Science Basis - Anthropogenic and Natural Radiative Forcing Supplementary Material" (PDF). Cambridge University Press.
- ↑ Eckburg P; Lepp P; Relman D (2003). "Archaea and their potential role in human disease". Infect Immun. 71 (2): 591–6. doi:10.1128/IAI.71.2.591-596.2003. PMC 145348
 . PMID 12540534.
. PMID 12540534. - ↑ Cavicchioli R; Curmi P; Saunders N; Thomas T (2003). "Pathogenic archaea: do they exist?". BioEssays. 25 (11): 1119–28. doi:10.1002/bies.10354. PMID 14579252.
- ↑ Lepp P; Brinig M; Ouverney C; Palm K; Armitage G; Relman D (2004). "Methanogenic Archaea and human periodontal disease". Proceedings of the National Academy of Sciences of the United States of America. 101 (16): 6176–81. Bibcode:2004PNAS..101.6176L. doi:10.1073/pnas.0308766101. PMC 395942
 . PMID 15067114.
. PMID 15067114. - ↑ Vianna ME; Conrads G; Gomes BP; Horz HP (April 2006). "Identification and quantification of archaea involved in primary endodontic infections". J. Clin. Microbiol. 44 (4): 1274–82. doi:10.1128/JCM.44.4.1274-1282.2006. PMC 1448633
 . PMID 16597851.
. PMID 16597851. - ↑ Waters E; et al. (October 2003). "The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism". Proceedings of the National Academy of Sciences of the United States of America. 100 (22): 12984–8. Bibcode:2003PNAS..10012984W. doi:10.1073/pnas.1735403100. PMC 240731
 . PMID 14566062.
. PMID 14566062. - ↑ Jahn U; Gallenberger M; Paper W; et al. (March 2008). "Nanoarchaeum equitans and Ignicoccus hospitalis: new insights into a unique, intimate association of two archaea". J. Bacteriol. 190 (5): 1743–50. doi:10.1128/JB.01731-07. PMC 2258681
 . PMID 18165302.
. PMID 18165302. - ↑ Baker BJ; Comolli LR; Dick GJ; Hauser LJ; Hyatt D; Dill BD; Land ML; VerBerkmoes NC; Hettich RL; Banfield JF (May 2010). "Enigmatic, ultrasmall, uncultivated Archaeaa". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8806–8811. Bibcode:2010PNAS..107.8806B. doi:10.1073/pnas.0914470107. PMC 2889320
 . PMID 20421484.
. PMID 20421484. - ↑ Chaban B; Ng SY; Jarrell KF (February 2006). "Archaeal habitats—from the extreme to the ordinary". Can. J. Microbiol. 52 (2): 73–116. doi:10.1139/w05-147. PMID 16541146.
- ↑ Schink B (June 1997). "Energetics of syntrophic cooperation in methanogenic degradation". Microbiol. Mol. Biol. Rev. 61 (2): 262–80. PMC 232610
 . PMID 9184013.
. PMID 9184013. - ↑ Lange, M; Westermann, P; Ahring, BK (2005). "Archaea in protozoa and metazoa". Applied Microbiology and Biotechnology. 66 (5): 465–474. doi:10.1007/s00253-004-1790-4. PMID 15630514.
- ↑ van Hoek AH; van Alen TA; Sprakel VS; et al. (1 February 2000). "Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates". Mol. Biol. Evol. 17 (2): 251–8. doi:10.1093/oxfordjournals.molbev.a026304. PMID 10677847.
- ↑ Preston, C.M; Wu, K.Y; Molinski, T.F; Delong, E.F (1996). "A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov". Proceedings of the National Academy of Sciences of the United States of America. 93 (13): 6241–6. Bibcode:1996PNAS...93.6241P. doi:10.1073/pnas.93.13.6241. PMC 39006
 . PMID 8692799.
. PMID 8692799. - ↑ Eckburg PB; et al. (June 2005). "Diversity of the human intestinal microbial flora". Science. 308 (5728): 1635–8. Bibcode:2005Sci...308.1635E. doi:10.1126/science.1110591. PMC 1395357
 . PMID 15831718.
. PMID 15831718. - ↑ Samuel BS; Gordon JI (June 2006). "A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism". Proceedings of the National Academy of Sciences of the United States of America. 103 (26): 10011–6. Bibcode:2006PNAS..10310011S. doi:10.1073/pnas.0602187103. PMC 1479766
 . PMID 16782812.
. PMID 16782812. - ↑ Wegley, L; Yu, Y; Breitbart, M; Casas, V; Kline, D.I; Rohwer, F (2004). "Coral-associated Archaea" (PDF). Marine Ecology Progress Series. 273: 89–96. doi:10.3354/meps273089. Archived from the original (PDF) on 11 September 2008.
- ↑ Chelius MK; Triplett EW (April 2001). "The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L". Microb. Ecol. 41 (3): 252–63. doi:10.1007/s002480000087. JSTOR 4251818. PMID 11391463.
- ↑ Simon HM; Dodsworth JA; Goodman RM (October 2000). "Crenarchaeota colonize terrestrial plant roots". Environ. Microbiol. 2 (5): 495–505. doi:10.1046/j.1462-2920.2000.00131.x. PMID 11233158.
- ↑ Breithaupt H (2001). "The hunt for living gold. The search for organisms in extreme environments yields useful enzymes for industry". EMBO Rep. 2 (11): 968–71. doi:10.1093/embo-reports/kve238. PMC 1084137
 . PMID 11713183.
. PMID 11713183. - 1 2 Egorova K; Antranikian G (2005). "Industrial relevance of thermophilic Archaea". Current Opinion in Microbiology. 8 (6): 649–55. doi:10.1016/j.mib.2005.10.015. PMID 16257257.
- ↑ Synowiecki J; Grzybowska B; Zdziebło A (2006). "Sources, properties and suitability of new thermostable enzymes in food processing". Crit Rev Food Sci Nutr. 46 (3): 197–205. doi:10.1080/10408690590957296. PMID 16527752.
- ↑ Jenney FE; Adams MW (January 2008). "The impact of extremophiles on structural genomics (and vice versa)". Extremophiles. 12 (1): 39–50. doi:10.1007/s00792-007-0087-9. PMID 17563834.
- ↑ Schiraldi C; Giuliano M; De Rosa M (2002). "Perspectives on biotechnological applications of archaea". Archaea. 1 (2): 75–86. doi:10.1155/2002/436561. PMC 2685559
 . PMID 15803645.
. PMID 15803645. - ↑ Norris PR; Burton NP; Foulis NA (2000). "Acidophiles in bioreactor mineral processing". Extremophiles. 4 (2): 71–6. doi:10.1007/s007920050139. PMID 10805560.
- ↑ Shand RF; Leyva KJ (2008). "Archaeal Antimicrobials: An Undiscovered Country". In Blum P. Archaea: New Models for Prokaryotic Biology. Caister Academic Press. ISBN 978-1-904455-27-1.
Further reading
- Howland, John L. (2000). The Surprising Archaea: Discovering Another Domain of Life. Oxford University. ISBN 978-0-19-511183-5.
- Martinko JM; Madigan MT (2005). Brock Biology of Microorganisms (11th ed.). Englewood Cliffs, N.J: Prentice Hall. ISBN 0-13-144329-1.
- Garrett RA; Klenk H (2005). Archaea: Evolution, Physiology and Molecular Biology. WileyBlackwell. ISBN 1-4051-4404-1.
- Cavicchioli R (2007). Archaea: Molecular and Cellular Biology. American Society for Microbiology. ISBN 1-55581-391-7.
- Blum P, ed. (2008). Archaea: New Models for Prokaryotic Biology. Caister Academic Press. ISBN 978-1-904455-27-1.
- Lipps G (2008). "Archaeal Plasmids". Plasmids: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-35-6.
- Sapp, Jan (2009). The New Foundations of Evolution. On the Tree of Life. New York: Oxford University Press. ISBN 0-19-538850-X.
- Schaechter, M (2009). Archaea (Overview) in The Desk Encyclopedia of Microbiology (2nd ed.). San Diego and London: Elsevier Academic Press. ISBN 978-0-12-374980-2.
External links
| Wikispecies has information related to: Archaea |
General
- Introduction to the Archaea, ecology, systematics and morphology
- Oceans of Archaea – E.F. DeLong, ASM News, 2003
Classification
- NCBI taxonomy page on Archaea
- Genera of the domain Archaea – list of Prokaryotic names with Standing in Nomenclature
- Shotgun sequencing finds nanoorganisms – discovery of the ARMAN group of archaea
Genomics
- Browse any completed archaeal genome at UCSC
- Comparative Analysis of Archaeal Genomes (at DOE's IMG system)