Cell surface receptor
.png)
Cell surface receptors (membrane receptors, transmembrane receptors) are receptors at the surface of a cell (built into its cell membrane) that act in cell signaling by receiving (binding to) extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the outside world. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane.
Structure and mechanism
Transmembrane receptors are typically classified based on their tertiary structure. But, if the three-dimensional structure is yet undiscovered, then they can be classified based on experimentally verifiable membrane topology. The simplest polypeptide chains are found to cross the lipid bilayer once; while others, such as the G-protein coupled receptors, cross as many as seven times. There are various kinds, such as glycoprotein and lipoprotein.[1] Hundreds of different receptors are known and many more have yet to be studied.[2][3] Many membrane receptors include transmembrane proteins. Each cell membrane can have several kinds of membrane receptor, in varying surface distribution. A specific receptor may also be differently distributed on different membrane surfaces, depending on the membrane sort and cell function. Since receptors usually cluster on the membrane surface,[4][5] the placement of every receptor on each membrane surface is heterogeneous.
Mechanism
Two current models have been proposed to explain transmembrane receptors mechanism, dimerization considered as current standard.
- Dimerization: This dimerization model suggests that prior to ligand binding, receptors exist in a monomeric form. When contact occurs with a ligand, receptors bind together to form a dimer, a functional compound receptor composed of two structural similar monomers.
- Rotation Model: Ligand binding to the extracellular part of the receptor induces the rotation of the receptor's transmembrane region inside the cell membrane, in doing so regulate it's activity inside the cell. Prior to ligand binding, the extracellular protein loses flexibility while the intracellular portion gains it.[6]
Domains
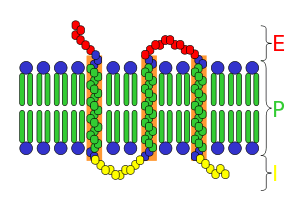
Like any integral membrane protein, a transmembrane receptor may be divided into three domains.
Extracellular domain
The extracellular domain juts externally from the cell or organelle. If the polypeptide chain crosses the bilayer several times, the external domain comprises loops entwined through the membrane. By definition, a receptor's main function is to recognize and respond to a type of ligand. For example, a neurotransmitter, hormone, or atomic ions may each bind to the extracellular domain as a ligand coupled to receptor. Klotho is an enzyme which effects a receptor to recognize the ligand (FGF23).
Transmembrane domain
In the majority of receptors with known structures, transmembrane alpha helices constitute most of the transmembrane component. In certain receptors, such as the nicotinic acetylcholine receptor, the transmembrane domain forms a protein pore through the membrane, or around the ion channel. Upon activation of an extracellular domain by binding of the appropriate ligand, the pore becomes accessible to ions, which then diffuse. In other receptors, the transmembrane domains undergo a conformational change upon binding, which effects intracellular conditions. In some receptors, such as members of the 7TM superfamily, the transmembrane domain includes a ligand binding pocket. Bacteriorhodopsin is an example, the detailed structure of which has been determined by crystallography.
Intracellular domain
The intracellular (or cytoplasmic) domain of the receptor interacts with the interior of the cell or organelle, relaying the signal. There are two fundamental paths for this interaction:
- The intracellular domain communicates via protein-protein interactions against effector proteins, which in turn pass a signal to the destination.
- With enzyme-linked receptors, the intracellular domain has enzymatic activity. Often, this is tyrosine kinase activity. The enzymatic activity can also be due to an enzyme associated with the intracellular domain.
Signal transduction

Signal transduction processes through membrane receptors involve the external reactions, in which the ligand binds to a membrane receptor, and the internal reactions, in which intracellular response is triggered.[7][8]
Signal transduction through membrane receptors requires four parts:
- Extracellular signaling molecule: an extracellular signaling molecule is produced by one cell and is at least capable of traveling to neighboring cells.
- Receptor protein: cells must have cell surface receptor proteins which bind to the signaling molecule and communicate inward into the cell.
- Intracellular signaling proteins: these pass the signal to the organelles of the cell. Binding of the signal molecule to the receptor protein will activate intracellular signaling proteins that initiate a signaling cascade.
- Target proteins: the conformations or other properties of the target proteins are altered when a signaling pathway is active and changes the behavior of the cell.[8]

Membrane receptors are mainly divided by structure and function into 3 classes: The ion channel-linked receptor; The enzyme-linked receptor; and The G protein-coupled receptor.
- Ion channel linked receptors have ion channels for anions and cations, and constitute a large family of multipass transmembrane proteins. They participate in rapid signaling events usually found in electrically active cells such as neurons. They are also called ligand-gated ion channels. Opening and closing of ion channels is controlled by neurotransmitters.
- Enzyme-linked receptors are either enzymes themselves, or are directly activate associated enzymes. These are typically single-pass transmembrane receptors, with the enzymatic component of the receptor kept intracellular. The majority of enzyme-linked receptors are, or associate with, protein kinases.
- G protein-coupled receptors are integral membrane proteins that possess seven transmembrane helices. These receptors activate a G protein with ligand binding. G-protein is a trimeric protein. The 3 subunits are called α、β and γ. The α subunit can bind with guanosine diphosphate, GDP. This causes phosphorylation of the GDP to guanosine triphosphate, GTP, and activates the α subunit to then dissociate from the β and γ subunits. The activated α subunit can further effect intracellular signaling proteins or target functional proteins directly.
Ion channel-linked receptor
During the signal transduction event in a neuron, the neurotransmitter binds to the receptor and alters the conformation of the protein. This opens the ion channel, allowing extracellular ions into the cell. Ion permeability of the plasma membrane is altered, and this transforms the extracellular chemical signal into an intracellular electric signal which alters the cell excitability.[9]
Acetylcholine receptor is a receptor linked to a cation channel. The protein consists of 4 subunits: α, β, γ, and δ subunits. There are two α subunits, with one acetylcholine binding site each. This receptor can exist in three conformations. The closed and unoccupied state is the native protein conformation. As two molecules of acetylcholine both bind to the binding sites on α subunits, the conformation of the receptor is altered and the gate is opened, allowing for the entry of many ions and small molecules. However, this open and occupied state only lasts for a minor duration and then the gate is closed, becoming the closed and occupied state. The two molecules of acetylcholine will soon dissociate from the receptor, returning it to the native closed and unoccupied state.[10][11]
Enzyme-linked receptors
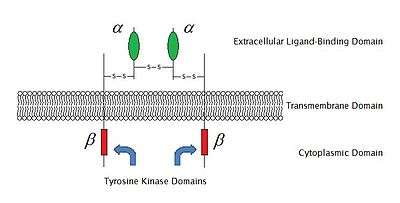
As of 2009, there are 6 known types of enzyme-linked receptors: Receptor tyrosine kinases; Tyrosine kinase associated receptors; Receptor-like tyrosine phosphatases; Receptor serine/threonine kinases; Receptor Guanylyl cyclases and Histidine kinase associated receptors. Receptor tyrosine kinases have the largest population and widest application. The majority of these molecules are receptors for growth factors and hormones like epidermal growth factor (EGF), platelet derived growth factor (PDGF), fibroblast growth factor (FGF), hepatocyte growth factor (HGF), insulin, nerve growth factor (NGF) etc.
Most of these receptors will dimerize after binding with their ligands, in order to activate further signal transductions. For example, after the epidermal growth factor (EGF) receptor binds with its ligand EGF, the two receptors dimerize and then undergo phosphorylation of the tyrosine residues in the enzyme portion of each receptor molecule. This will activate the tyrosine protein kinase and catalyze further intracellular reactions.
G protein-coupled receptors
G protein-coupled receptors comprise a large protein family of transmembrane receptors. They are found only in eukaryotes.[12] The ligands which bind and activate these receptors include: photosensitive compounds, odors, pheromones, hormones, and neurotransmitters. These vary in size from small molecules to peptides and large proteins. G protein-coupled receptors are involved in many diseases, and thus are the targets of many modern medicinal drugs.[13]
There are two principal signal transduction pathways involving the G-protein coupled receptors: cAMP signal pathway and Phosphatidylinositol signal pathway.[14] Both activate a G protein ligand binding. G-protein is a trimeric protein. The 3 subunits are called α、β and γ. The α subunit can bind with guanosine diphosphate, GDP. This causes phosphorylation of the GDP to guanosine triphosphate, GTP, and activates the α subunit, which then dissociates from the β and γ subunits. The activated α subunit can further affect intracellular signaling proteins or target functional proteins directly.
Membrane receptor-related disease
If the membrane receptors are denatured or deficient, the signal transduction can be hindered and cause diseases. Some diseases are caused by disorders of membrane receptor function. This is due to deficiency or degradation of the receptor via changes in the genes that encode and regulate the receptor protein. The membrane receptor TM4SF5 influences the migration of hepatic cells and hepatoma.[15] Also, the cortical NMDA receptor influences membrane fluidity, and is altered in Alzheimer's disease.[16] When the cell is infected by a non-enveloped virus, the virus first binds to specific membrane receptors and then passes itself or a subviral component to the cytoplasmic side of the cellular membrane. In the case of poliovirus, it is known in vitro that interactions with receptors cause conformational rearrangements which release a virion protein called VP4.The N terminus of VP4 is myristylated and thus hydrophobic【myristic acid=CH3(CH2)12COOH】. It is proposed that the conformational changes induced by receptor binding result in the attachment of myristic acid on VP4 and the formation of a channel for RNA.
Structure-based drug design
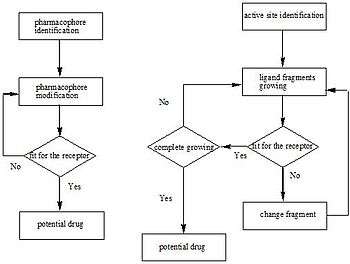
Through methods such as X-ray crystallography and NMR spectroscopy, the information about 3D structures of target molecules has increased dramatically, and so has structural information about the ligands. This drives rapid development of structure-based drug design. Some of these new drugs target membrane receptors. Current approaches to structure-based drug design can be divided into two categories. The first category is about determining ligands for a given receptor. This is usually accomplished through database queries, biophysical simulations, and the construction of chemical libraries. In each case, a large number of potential ligand molecules are screened to find those fitting the binding pocket of the receptor. This approach is usually referred to as ligand-based drug design. The key advantage of searching a database is that it saves time and power to obtain new effective compounds. Another approach of structure-based drug design is about combinatorially mapping ligands, which is referred to as receptor-based drug design. In this case, ligand molecules are engineered within the constraints of a binding pocket by assembling small pieces in a stepwise manner. These pieces can be either atoms or molecules. The key advantage of such a method is that novel structures can be discovered.[17][18][19]
See also
Other examples
- Adrenergic receptor,
- Olfactory receptors,
- Receptor tyrosine kinases
- Epidermal growth factor receptor
- Insulin Receptor
- Fibroblast growth factor receptors,
- High affinity neurotrophin receptors
- Eph Receptors
- Integrins
- Low Affinity Nerve Growth Factor Receptor
- NMDA receptor
- Several Immune receptors
References
- ↑ Cuatrecasas P. (1974). "MEMBRANE RECEPTORS". Annual Review of Biochemistry. 43 (0): 169–214. doi:10.1146/annurev.bi.43.070174.001125. PMID 4368906.
- ↑ Dautzenberg FM, Hauger RL (February 2002). "The CRF peptide family and their receptors: yet more partners discovered". Trends Pharmacol. Sci. 23 (2): 71–7. doi:10.1016/S0165-6147(02)01946-6. PMID 11830263.
- ↑ Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I (May 2009). "Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors". Nature. 459 (7246): 574–7. doi:10.1038/nature08029. PMID 19387439.
- ↑ Rothberg K.G.; Ying Y.S.; Kamen B.A.; Anderson R.G. (1990). "Cholesterol controls the clustering of the glycophospholipid-anchored membrane receptor for 5-methyltetrahydrofolate". The Journal of Cell Biology. 111 (6): 2931–2938. doi:10.1083/jcb.111.6.2931. PMC 2116385
 . PMID 2148564.
. PMID 2148564. - ↑ Jacobson C.; Côté P.D.; Rossi S.G.; Rotundo R.L.; Carbonetto S. (2001). "The Dystroglycan Complex Is Necessary for Stabilization of Acetylcholine Receptor Clusters at Neuromuscular Junctions and Formation of the Synaptic Basement Membrane". The Journal of Cell Biology. 152 (3): 435–450. doi:10.1083/jcb.152.3.435. PMC 2195998
 . PMID 11157973.
. PMID 11157973. - ↑ Maruyama, Ichiro N. (2015-09-01). "Activation of transmembrane cell-surface receptors via a common mechanism? The "rotation model"". BioEssays. 37 (9): 959–967. doi:10.1002/bies.201500041. ISSN 1521-1878.
- ↑ Ullricha A.,Schlessingerb J.; Schlessinger, J (1990). "Signal transduction by receptors with tyrosine kinase activity". Cell. 61 (2): 203–212. doi:10.1016/0092-8674(90)90801-K. PMID 2158859.
- 1 2 Kenneth B. Storey (1990). Functional Metabolism. Wiley-IEEE. pp. 87–94. ISBN 0-471-41090-X.
- ↑ Hille B. (2001). Ion channels of excitable membranes. Sunderland, Mass. ISBN 0-87893-321-2.
- ↑ Miyazawa A.; Fujiyoshi Y.; Unwin N. (2003). "Structure and gating mechanism of the acetylcholine receptor pore". Nature. 423 (6943): 949–955. doi:10.1038/nature01748. PMID 12827192.
- ↑ Akabas M.H.; Stauffer D.A.; Xu M.; Karlin A. (1992). "Acetylcholine receptor channel structure probed in cysteine-substitution mutants". Science. 258 (5080): 307–310. doi:10.1126/science.1384130. PMID 1384130.
- ↑ King N, Hittinger CT, Carroll SB (2003). "Evolution of key cell signaling and adhesion protein families predates animal origins". Science. 301 (5631): 361–3. doi:10.1126/science.1083853. PMID 12869759.
- ↑ Filmore, David (2004). "It's a GPCR world". Modern Drug Discovery. American Chemical Society. 2004 (November): 24–28.
- ↑ Gilman A.G. (1987). "G Proteins: Transducers of Receptor-Generated Signals". Annual Review of Biochemistry. 56: 615–649. doi:10.1146/annurev.bi.56.070187.003151. PMID 3113327.
- ↑ Müller-Pillascha F.; Wallrappa C.; Lachera U.; Friessb H.; Büchlerb M.; Adlera G.; Gress T. M. (1998). "Identification of a new tumour-associated antigen TM4SF5 and its expression in human cancer". Gene. 208 (1): 25–30. doi:10.1016/S0378-1119(97)00633-1. PMID 9479038.
- ↑ Scheuer K.; Marasb A.; Gattazb W.F.; Cairnsc N.; Förstlb H.; Müller W.E. (1996). "Cortical NMDA Receptor Properties and Membrane Fluidity Are Altered in Alzheimer's Disease". Dementia. 7 (4): 210–214. doi:10.1159/000106881. PMID 8835885.
- ↑ Wang R.; Gao Y.; Lai L. (2000). "LigBuilder: A Multi-Purpose Program for Structure-Based Drug Design". Journal of Molecular Modeling. 6 (7–8): 498–516. doi:10.1007/s0089400060498.
- ↑ Schneider G.; Fechner U. (2005). "Computer-based de novo design of drug-like molecules". Nature Reviews Drug Discovery. 4 (8): 649–663. doi:10.1038/nrd1799. PMID 16056391.
- ↑ Jorgensen W.L. (2004). "The Many Roles of Computation in Drug Discovery". Science. 303 (5665): 1813–1818. doi:10.1126/science.1096361. PMID 15031495.
External links
- IUPHAR GPCR Database
- Cell Surface Receptors at the US National Library of Medicine Medical Subject Headings (MeSH)